Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes.
Xu, Q., Traag, B.A., Willemse, J., McMullan, D., Miller, M.D., Elsliger, M.A., Abdubek, P., Astakhova, T., Axelrod, H.L., Bakolitsa, C., Carlton, D., Chen, C., Chiu, H.J., Chruszcz, M., Clayton, T., Das, D., Deller, M.C., Duan, L., Ellrott, K., Ernst, D., Farr, C.L., Feuerhelm, J., Grant, J.C., Grzechnik, A., Grzechnik, S.K., Han, G.W., Jaroszewski, L., Jin, K.K., Klock, H.E., Knuth, M.W., Kozbial, P., Krishna, S.S., Kumar, A., Marciano, D., Minor, W., Mommaas, A.M., Morse, A.T., Nigoghossian, E., Nopakun, A., Okach, L., Oommachen, S., Paulsen, J., Puckett, C., Reyes, R., Rife, C.L., Sefcovic, N., Tien, H.J., Trame, C.B., van den Bedem, H., Wang, S., Weekes, D., Hodgson, K.O., Wooley, J., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A., van Wezel, G.P.(2009) J Biol Chem 284: 25268-25279
- PubMed: 19567872
- DOI: https://doi.org/10.1074/jbc.M109.018564
- Primary Citation of Related Structures:
3CM1 - PubMed Abstract:
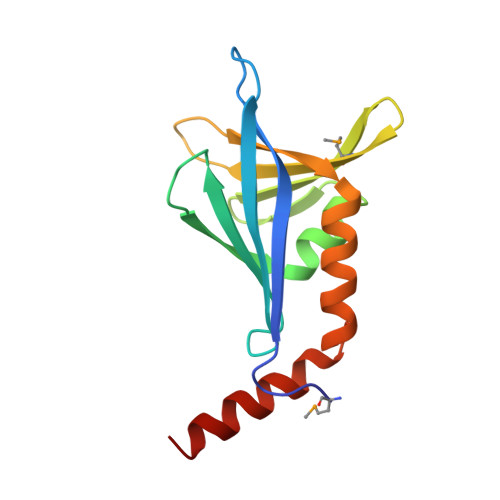
SsgA-like proteins (SALPs) are a family of homologous cell division-related proteins that occur exclusively in morphologically complex actinomycetes. We show that SsgB, a subfamily of SALPs, is the archetypal SALP that is functionally conserved in all sporulating actinomycetes. Sporulation-specific cell division of Streptomyces coelicolor ssgB mutants is restored by introduction of distant ssgB orthologues from other actinomycetes. Interestingly, the number of septa (and spores) of the complemented null mutants is dictated by the specific ssgB orthologue that is expressed. The crystal structure of the SsgB from Thermobifida fusca was determined at 2.6 A resolution and represents the first structure for this family. The structure revealed similarities to a class of eukaryotic "whirly" single-stranded DNA/RNA-binding proteins. However, the electro-negative surface of the SALPs suggests that neither SsgB nor any of the other SALPs are likely to interact with nucleotide substrates. Instead, we show that a conserved hydrophobic surface is likely to be important for SALP function and suggest that proteins are the likely binding partners.
Organizational Affiliation:
Joint Center for Structural Genomics, SLAC National Accelerator Laboratory, Menlo Park, California 94025, USA.