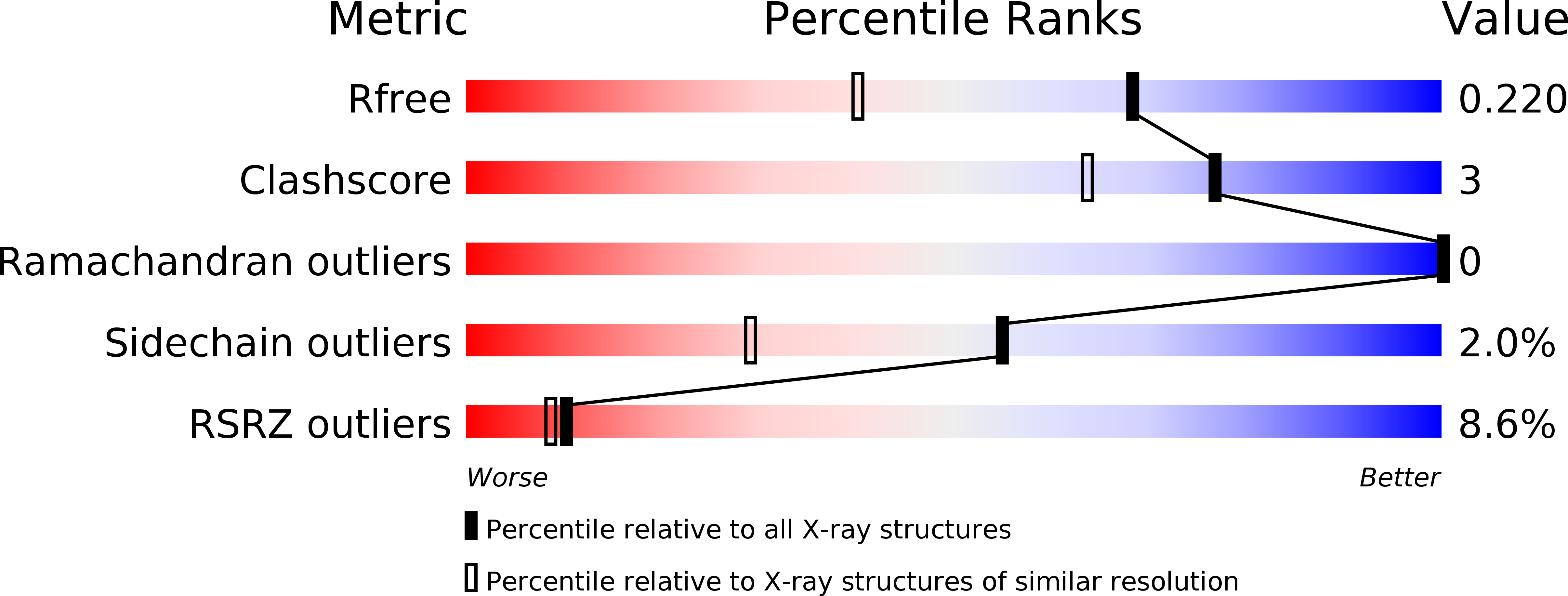
Crystal structure of textilinin-1, a Kunitz-type serine protease inhibitor from the venom of the Australian common brown snake (Pseudonaja textilis).
Millers, E.K., Trabi, M., Masci, P.P., Lavin, M.F., de Jersey, J., Guddat, L.W.(2009) FEBS J 276: 3163-3175
- PubMed: 19490116
- DOI: https://doi.org/10.1111/j.1742-4658.2009.07034.x
- Primary Citation of Related Structures:
3BYB - PubMed Abstract:
Textilinin-1 is a Kunitz-type serine protease inhibitor isolated from the venom of the Australian common brown snake, Pseudonaja textilis. This molecule binds to and blocks the activity of a range of serine proteases, including plasmin and trypsin. Textilinin-1's ability to inhibit plasmin, a protease involved in fibrinolysis, has raised the possibility that it could be used as an alternative to aprotinin (Trasylol) as a systemic antibleeding agent in surgery. Here, the crystal structure of free recombinant textilinin-1 has been determined to 1.63 A, with three molecules observed in the asymmetric unit. All of these have a similar overall fold to aprotinin, except that the canonical loop for one of the molecules is inverted such that the side chain of the P1' residue, Val18, is partially buried by intramolecular contacts to Pro15, Thr13, and Ile36. In aprotinin, the P1' residue is Ala16, whose side chain is too small to form similar contacts. The loop inversion in textilinin-1 is facilitated by changes in backbone dihedral angles for the P1 and P2' residues, such that they alternate between values in the beta-sheet and alpha-helical regions of the Ramachandran plot. In a comparison with the structures of all other known Kunitz-type serine protease inhibitors, no such conformational variability has been observed. The presence of the bulkier valine as the P1' residue in textilinin-1 appears to be a major contributor to reducing the binding affinity for plasmin as compared to aprotinin (3.5 nm versus 0.053 nm) and could also account for an observed narrower binding specificity.
Organizational Affiliation:
School of Molecular and Microbial Sciences, The University of Queensland, Brisbane, Australia.