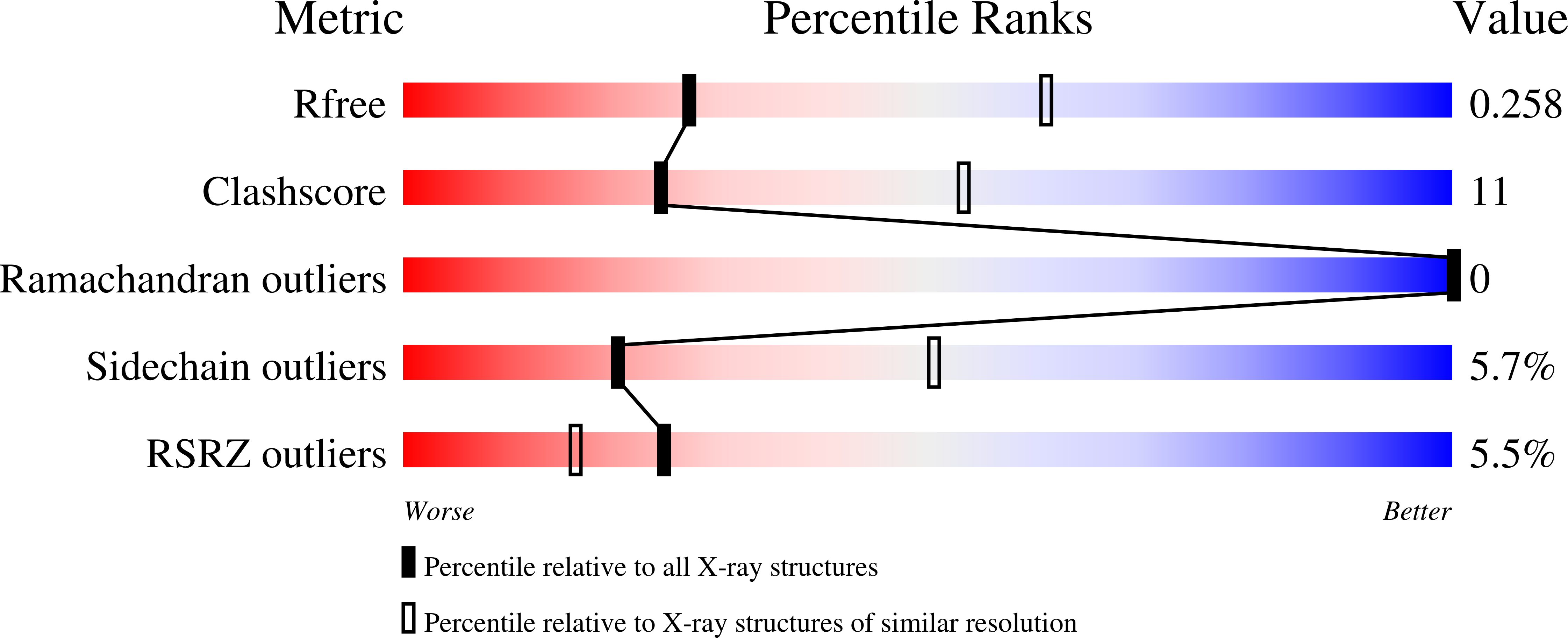
Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy.
Fujioka, Y., Noda, N.N., Nakatogawa, H., Ohsumi, Y., Inagaki, F.(2010) J Biol Chem 285: 1508-1515
- PubMed: 19889643
- DOI: https://doi.org/10.1074/jbc.M109.053520
- Primary Citation of Related Structures:
3A7O, 3A7P - PubMed Abstract:
Atg16 interacts with the Atg12-Atg5 protein conjugate through its N-terminal domain and self-assembles through its coiled-coil domain (CCD). Formation of the Atg12-Atg5.Atg16 complex is essential for autophagy, the bulk degradation process conserved among most eukaryotes. Here, we report the crystal structures of full-length Saccharomyces cerevisiae Atg16 at 2.8 A resolution and its CCD at 2.5 A resolution. The CCD and full-length Atg16 each exhibit an extended alpha-helix, 90 and 130 A, respectively, and form a parallel coiled-coil dimer in the crystals. Although the apparent molecular weight of Atg16 observed by gel-filtration chromatography suggests that Atg16 is tetrameric, an analytical ultracentrifugation study showed Atg16 as a dimer in solution, consistent with the crystal structure. Evolutionary conserved surface residues clustered at the C-terminal half of Atg16 CCD were shown to be crucial for autophagy. These results will give a structural basis for understanding the molecular functions and significance of Atg16 in autophagy.
Organizational Affiliation:
Department of Structural Biology, Graduate School of Pharmaceutical Sciences, Hokkaido University, Sapporo 060-0812, Japan.