Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
Otani, J., Arita, K., Kato, T., Kinoshita, M., Kimura, H., Suetake, I., Tajima, S., Ariyoshi, M., Shirakawa, M.(2013) J Biol Chem 288: 6351-6362
- PubMed: 23316048
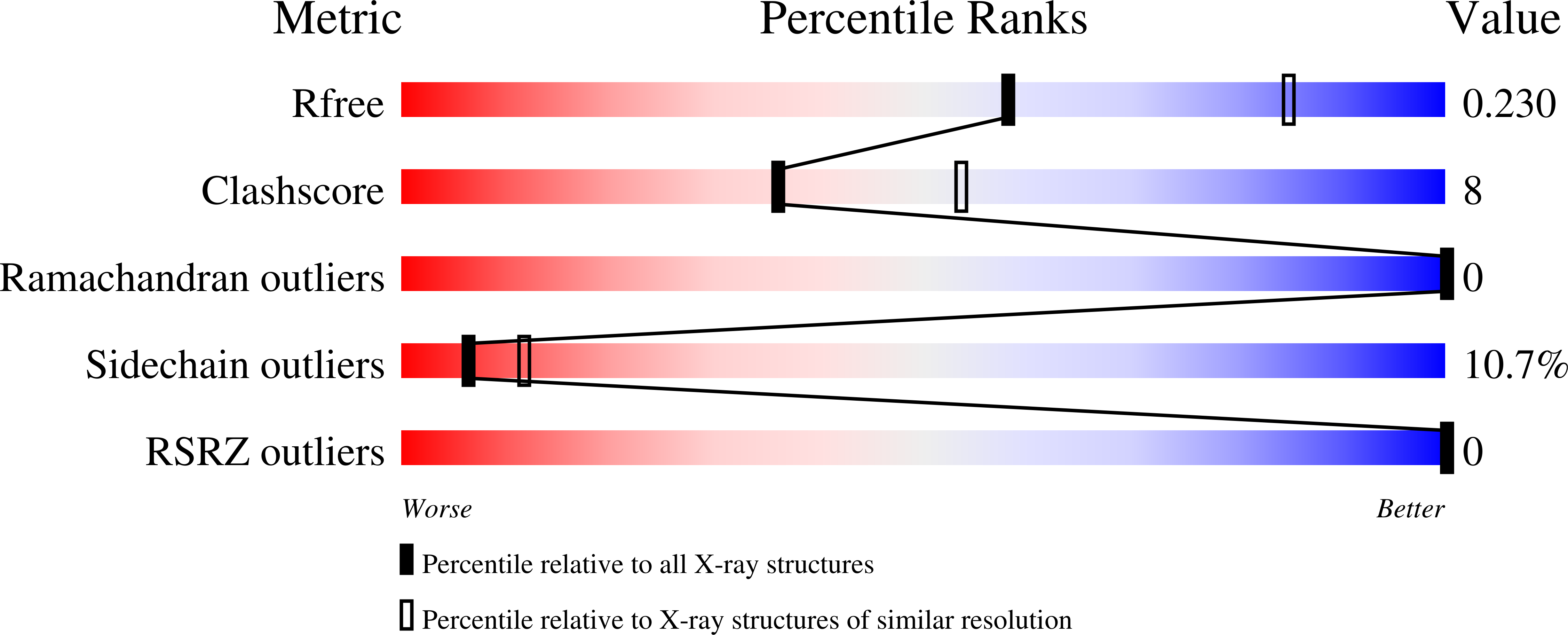
- DOI: https://doi.org/10.1074/jbc.M112.431098
- Primary Citation of Related Structures:
3VXV, 3VXX, 3VYB, 3VYQ - PubMed Abstract:
The methyl-CpG binding domain (MBD) protein MBD4 participates in DNA repair as a glycosylase that excises mismatched thymine bases in CpG sites and also functions in transcriptional repression. Unlike other MBD proteins, MBD4 recognizes not only methylated CpG dinucleotides ((5m)CG/(5m)CG) but also T/G mismatched sites generated by spontaneous deamination of 5-methylcytosine ((5m)CG/TG). The glycosylase activity of MBD4 is also implicated in active DNA demethylation initiated by the deaminase-catalyzed conversion of 5-methylcytosine to thymine. Here, we report the crystal structures of the MBD of MBD4 (MBDMBD4) complexed with (5m)CG/(5m)CG and (5m)CG/TG. The crystal structures show that the DNA interface of MBD4 has flexible structural features and harbors an extensive water network that supports its dual base specificities. Combined with the results of biochemical analyses, the crystal structure of MBD4 bound to 5-hydroxymethylcytosine further demonstrates that MBDMBD4 is able to recognize a wide range of 5-methylcytosine modifications through the unique water network. The versatile base recognition ability of MBDMBD4 implies multifunctional roles for MBD4 in the regulation of dynamic DNA methylation patterns coupled with deamination and/or oxidation of 5-methylcytosine.
Organizational Affiliation:
Graduate School of Engineering, Kyoto University, Kyoto 615-8510, Japan.