A new protein complex promoting the assembly of Rad51 filaments
Sasanuma, H., Tawaramoto, M.S., Lao, J.P., Hosaka, H., Sanda, E., Suzuki, M., Yamashita, E., Hunter, N., Shinohara, M., Nakagawa, A., Shinohara, A.(2013) Nat Commun 4: 1676-1676
- PubMed: 23575680
- DOI: https://doi.org/10.1038/ncomms2678
- Primary Citation of Related Structures:
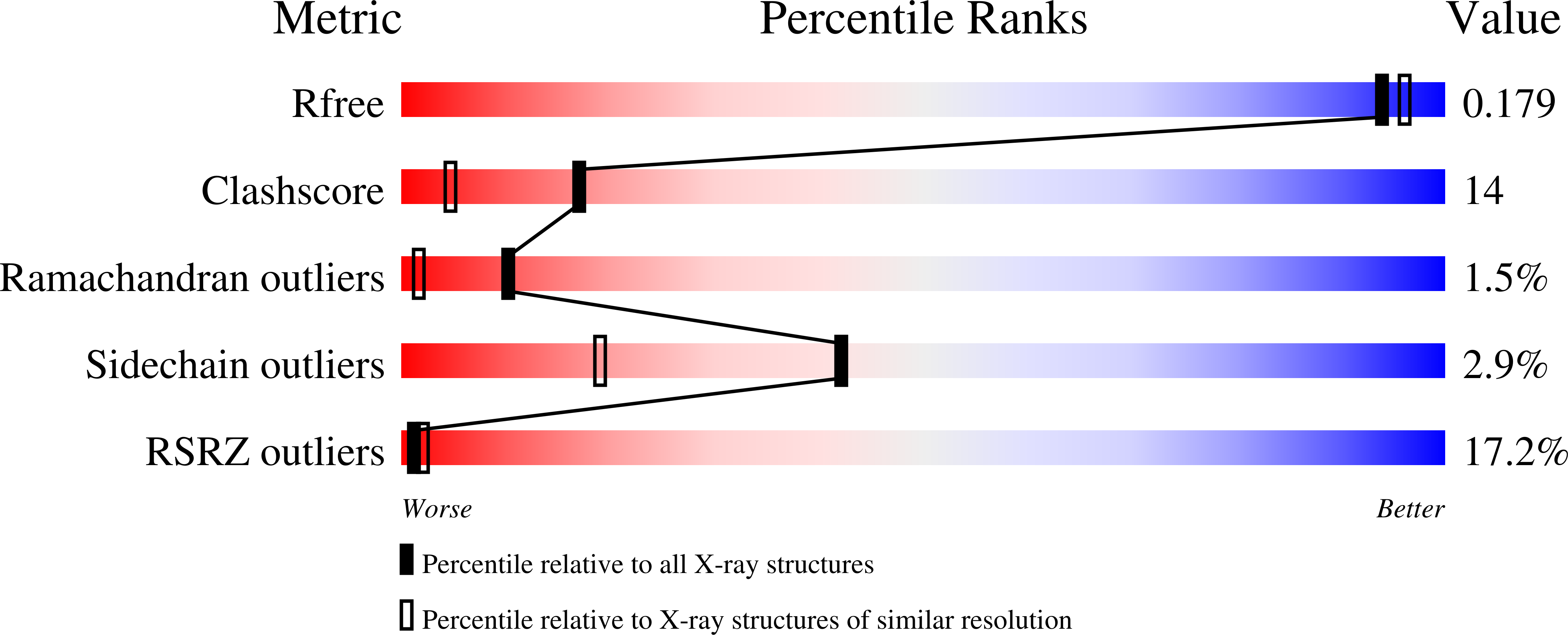
3VU9 - PubMed Abstract:
During homologous recombination, eukaryotic RecA homologue Rad51 assembles into a nucleoprotein filament on single-stranded DNA to catalyse homologous pairing and DNA-strand exchange with a homologous template. Rad51 nucleoprotein filaments are highly dynamic and regulated via the coordinated actions of various accessory proteins including Rad51 mediators. Here, we identify a new Rad51 mediator complex. The PCSS complex, comprising budding yeast Psy3, Csm2, Shu1 and Shu2 proteins, binds to recombination sites and is required for Rad51 assembly and function during meiosis. Within the hetero-tetramer, Psy3-Csm2 constitutes a core sub-complex with DNA-binding activity. In vitro, purified Psy3-Csm2 stabilizes the Rad51-single-stranded DNA complex independently of nucleotide cofactor. The mechanism of Rad51 stabilization is inferred by our high-resolution crystal structure, which reveals Psy3-Csm2 to be a structural mimic of the Rad51-dimer, a fundamental unit of the Rad51-filament. Together, these results reveal a novel molecular mechanism for this class of Rad51-mediators, which includes the human Rad51 paralogues.
Organizational Affiliation:
Institute for Protein Research, Graduate School of Science, Osaka University, Suita 565-0871, Japan.