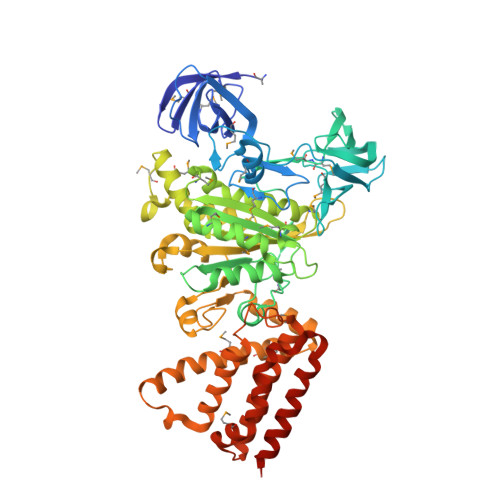
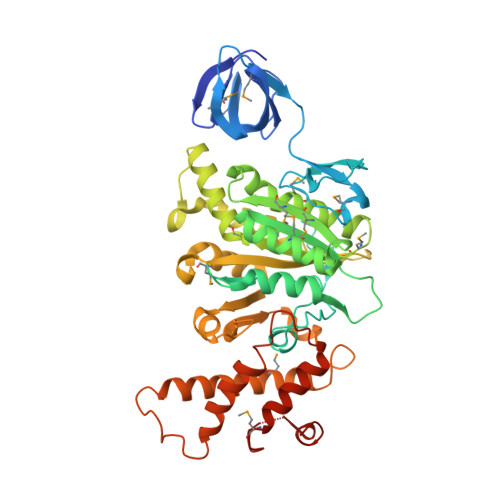
Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Arai, S., Saijo, S., Suzuki, K., Mizutani, K., Kakinuma, Y., Ishizuka-Katsura, Y., Ohsawa, N., Terada, T., Shirouzu, M., Yokoyama, S., Iwata, S., Yamato, I., Murata, T.(2013) Nature 493: 703-707
- PubMed: 23334411
- DOI: https://doi.org/10.1038/nature11778
- Primary Citation of Related Structures:
3VR2, 3VR3, 3VR4, 3VR5, 3VR6 - PubMed Abstract:
In various cellular membrane systems, vacuolar ATPases (V-ATPases) function as proton pumps, which are involved in many processes such as bone resorption and cancer metastasis, and these membrane proteins represent attractive drug targets for osteoporosis and cancer. The hydrophilic V(1) portion is known as a rotary motor, in which a central axis DF complex rotates inside a hexagonally arranged catalytic A(3)B(3) complex using ATP hydrolysis energy, but the molecular mechanism is not well defined owing to a lack of high-resolution structural information. We previously reported on the in vitro expression, purification and reconstitution of Enterococcus hirae V(1)-ATPase from the A(3)B(3) and DF complexes. Here we report the asymmetric structures of the nucleotide-free (2.8 Å) and nucleotide-bound (3.4 Å) A(3)B(3) complex that demonstrate conformational changes induced by nucleotide binding, suggesting a binding order in the right-handed rotational orientation in a cooperative manner. The crystal structures of the nucleotide-free (2.2 Å) and nucleotide-bound (2.7 Å) V(1)-ATPase are also reported. The more tightly packed nucleotide-binding site seems to be induced by DF binding, and ATP hydrolysis seems to be stimulated by the approach of a conserved arginine residue. To our knowledge, these asymmetric structures represent the first high-resolution view of the rotational mechanism of V(1)-ATPase.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Chiba University, 1-33 Yayoi-cho, Inage, Chiba 263-8522, Japan.