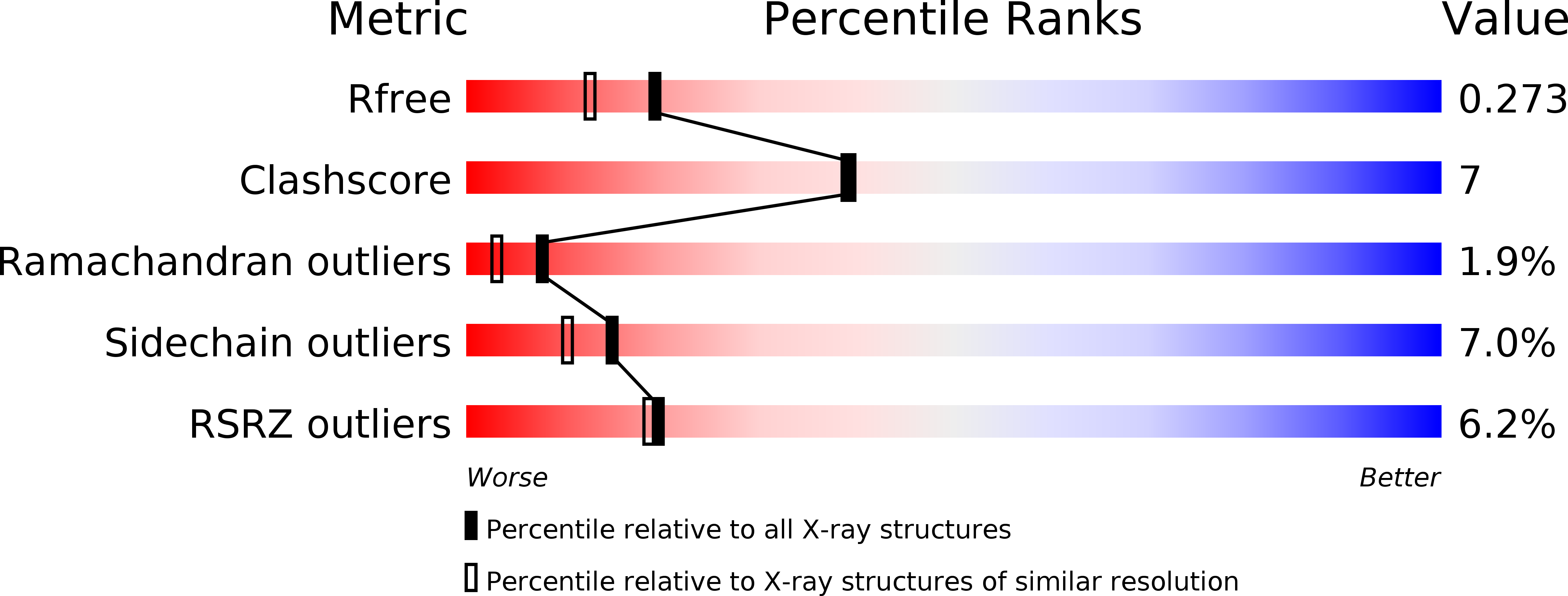
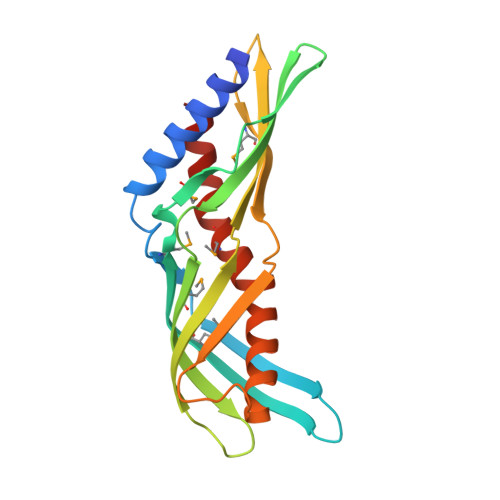
Crystal Structure of Der f 7, a Dust Mite Allergen from Dermatophagoides farinae.
Tan, K.W., Jobichen, C., Ong, T.C., Gao, Y.F., Tiong, Y.S., Wong, K.N., Chew, F.T., Sivaraman, J., Mok, Y.K.(2012) PLoS One 7: e44850-e44850
- PubMed: 22970319
- DOI: https://doi.org/10.1371/journal.pone.0044850
- Primary Citation of Related Structures:
3UV1 - PubMed Abstract:
Der f 7 is the group 7 allergen from the dust mite Dermatophagoides farinae, homologous to the major allergen Der p 7 from D. pteronyssinus. Monoclonal antibody that bind to residues Leu48 and Phe50 was found to inhibit IgE binding to residue Asp159, which is important for the cross-reactivity between Der f 7 and Der p 7. Here, we report the crystal structure of Der f 7 that shows an elongated and curved molecule consisting of two anti-parallel β-sheets--one 4-stranded and the other 5-stranded--that wrap around a long C-terminal helix. The overall fold of Der f 7 is similar to Der p 7 but key difference was found in the β1-β2 loop region. In Der f 7, Leu48 and Phe50 are in close proximity to Asp159, explaining why monoclonal antibody binding to Leu48 and Phe50 can inhibit IgE binding to Asp159. Both Der f 7 and Der p 7 bind weakly to polymyxin B via a similar binding site that is formed by the N-terminal helix, the 4-stranded β-sheet and the C-terminal helix. The thermal stability of Der f 7 is significantly lower than that of Der p 7, and the stabilities of both allergens are highly depend on pH. Der f 7 is homologous to Der p 7 in terms of the amino acid sequence and overall 3D structure but with significant differences in the region proximal to the IgE epitope and in thermal stability. The crystal structure of Der f 7 provides a basis for studying the function and allergenicity of this group of allergens.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore, Singapore, Singapore.