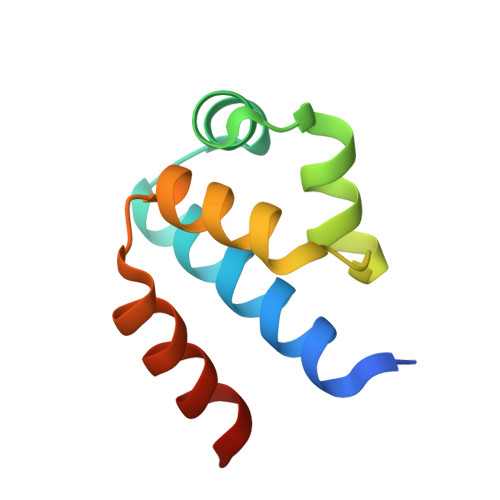
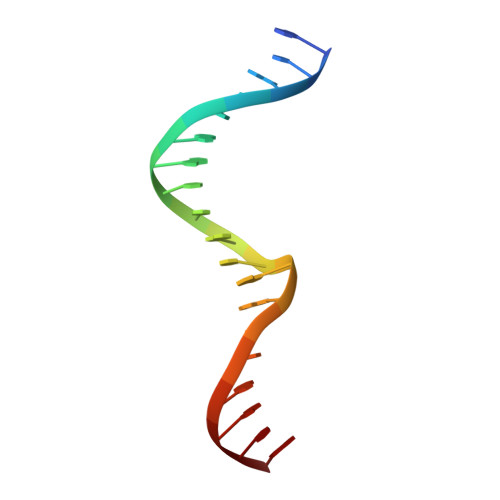
The structural basis of differential DNA sequence recognition by restriction-modification controller proteins.
Ball, N.J., McGeehan, J.E., Streeter, S.D., Thresh, S.J., Kneale, G.G.(2012) Nucleic Acids Res 40: 10532-10542
- PubMed: 22941636
- DOI: https://doi.org/10.1093/nar/gks718
- Primary Citation of Related Structures:
3UFD - PubMed Abstract:
Controller (C) proteins regulate the expression of restriction-modification (RM) genes in a wide variety of RM systems. However, the RM system Esp1396I is of particular interest as the C protein regulates both the restriction endonuclease (R) gene and the methyltransferase (M) gene. The mechanism of this finely tuned genetic switch depends on differential binding affinities for the promoters controlling the R and M genes, which in turn depends on differential DNA sequence recognition and the ability to recognize dual symmetries. We report here the crystal structure of the C protein bound to the M promoter, and compare the binding affinities for each operator sequence by surface plasmon resonance. Comparison of the structure of the transcriptional repression complex at the M promoter with that of the transcriptional activation complex at the R promoter shows how subtle changes in protein-DNA interactions, underpinned by small conformational changes in the protein, can explain the molecular basis of differential regulation of gene expression.
Organizational Affiliation:
Biomolecular Structure Group, Institute of Biomedical and Biomolecular Sciences, School of Biological Sciences, University of Portsmouth, Portsmouth PO1 2DY, UK.