B-DNA structure is intrinsically polymorphic: even at the level of base pair positions.
Maehigashi, T., Hsiao, C., Woods, K.K., Moulaei, T., Hud, N.V., Williams, L.D.(2012) Nucleic Acids Res 40: 3714-3722
- PubMed: 22180536
- DOI: https://doi.org/10.1093/nar/gkr1168
- Primary Citation of Related Structures:
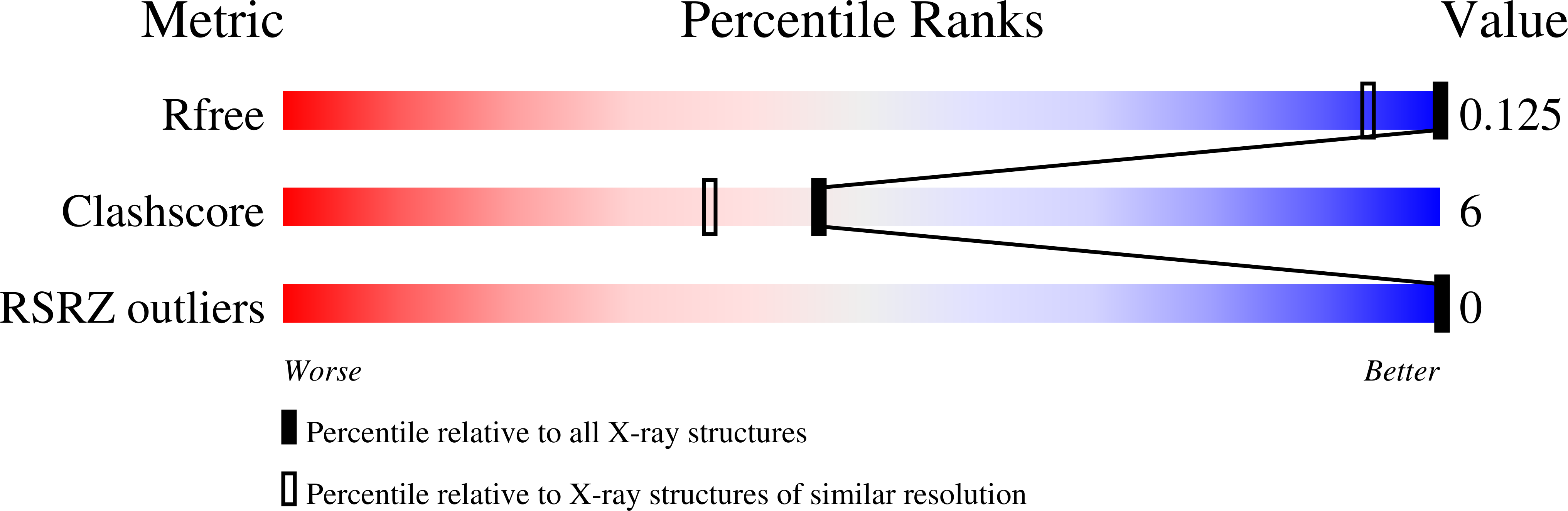
3U89 - PubMed Abstract:
Increasingly exact measurement of single crystal X-ray diffraction data offers detailed characterization of DNA conformation, hydration and electrostatics. However, instead of providing a more clear and unambiguous image of DNA, highly accurate diffraction data reveal polymorphism of the DNA atomic positions and conformation and hydration. Here we describe an accurate X-ray structure of B-DNA, painstakingly fit to a multistate model that contains multiple competing positions of most of the backbone and of entire base pairs. Two of ten base-pairs of CCAGGCCTGG are in multiple states distinguished primarily by differences in slide. Similarly, all the surrounding ions are seen to fractionally occupy discrete competing and overlapping sites. And finally, the vast majority of water molecules show strong evidence of multiple competing sites. Conventional resolution appears to give a false sense of homogeneity in conformation and interactions of DNA. In addition, conventional resolution yields an average structure that is not accurate, in that it is different from any of the multiple discrete structures observed at high resolution. Because base pair positional heterogeneity has not always been incorporated into model-building, even some high and ultrahigh-resolution structures of DNA do not indicate the full extent of conformational polymorphism.
Organizational Affiliation:
School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, GA 30332-0400, USA.