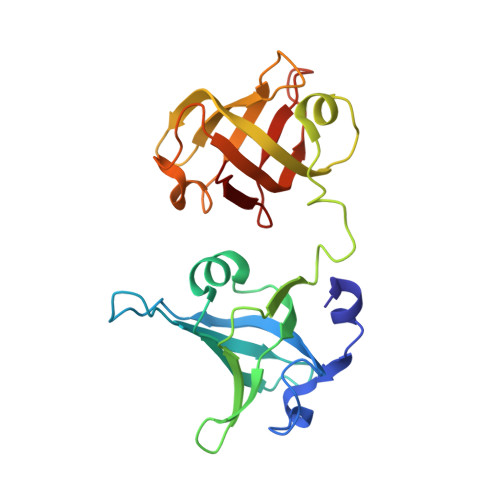
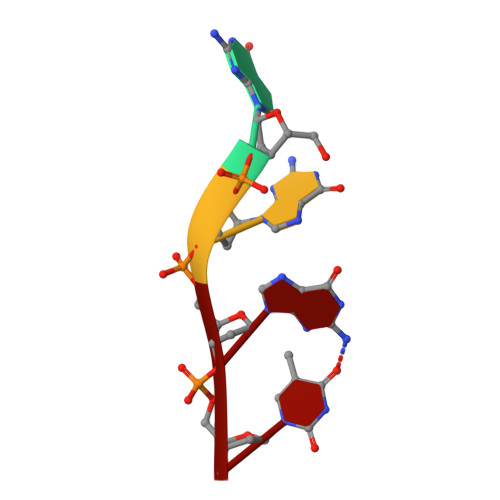
Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Zeng, Z., Min, B., Huang, J., Hong, K., Yang, Y., Collins, K., Lei, M.(2011) Proc Natl Acad Sci U S A 108: 20357-20361
- PubMed: 22143754
- DOI: https://doi.org/10.1073/pnas.1113624108
- Primary Citation of Related Structures:
3U4V, 3U4Z, 3U50, 3U58 - PubMed Abstract:
Telomerase copies its internal RNA template to synthesize telomeric DNA repeats. Unlike other polymerases, telomerase can retain its single-stranded product through multiple rounds of template dissociation and repositioning to accomplish repeat addition processivity (RAP). Tetrahymena telomerase holoenzyme RAP depends on a subunit, Teb1, with autonomous DNA-binding activity. Sequence homology and domain modeling suggest that Teb1 is a paralog of RPA70C, the largest subunit of the single-stranded DNA-binding factor replication protein (RPA), but unlike RPA, Teb1 binds DNA with high specificity for telomeric repeats. To understand the structural basis and significance of telomeric-repeat DNA recognition by Teb1, we solved crystal structures of three proposed Teb1 DNA-binding domains and defined amino acids of each domain that contribute to DNA interaction. Our studies indicate that two central Teb1 DNA-binding oligonucleotide/oligosaccharide-binding-fold domains, Teb1A and Teb1B, achieve high affinity and selectivity of telomeric-repeat recognition by principles similar to the telomere end-capping protein POT1 (protection of telomeres 1). An additional C-terminal Teb1 oligonucleotide/oligosaccharide-binding-fold domain, Teb1C, has features shared with the RPA70 C-terminal domain including a putative direct DNA-binding surface that is critical for high-RAP activity of reconstituted holoenzyme. The Teb1C zinc ribbon motif does not contribute to DNA binding but is nonetheless required for high-RAP activity, perhaps contributing to Teb1 physical association with the remainder of the holoenzyme. Our results suggest the biological model that high-affinity DNA binding by Teb1AB recruits holoenzyme to telomeres and subsequent Teb1C-DNA association traps product in a sliding-clamp-like manner that does not require high-affinity DNA binding for high stability of enzyme-product association.
Organizational Affiliation:
Howard Hughes Medical Institute, Department of Biological Chemistry, University of Michigan Medical School, Ann Arbor, MI 48109, USA.