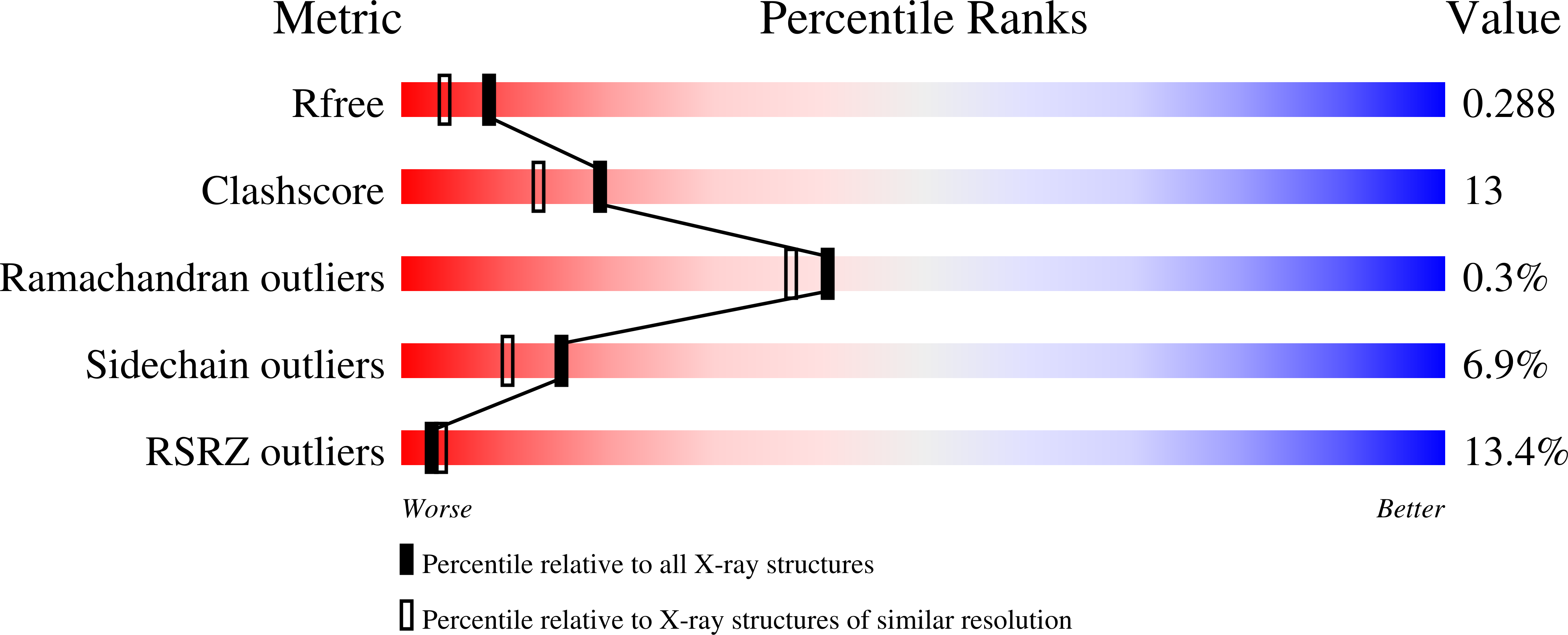
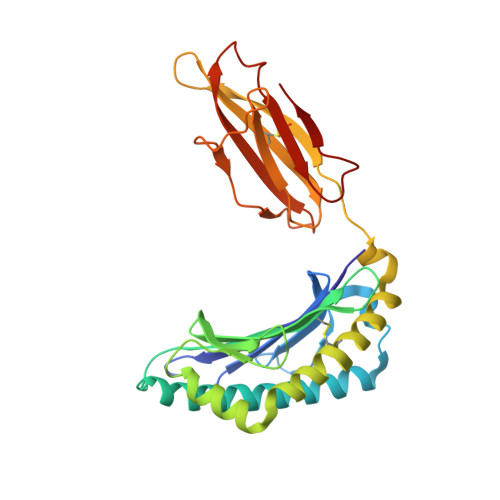
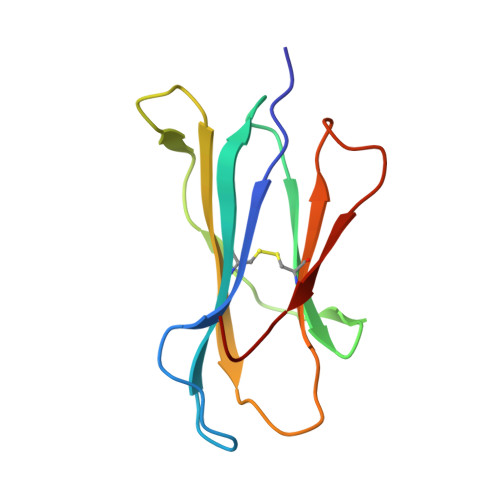
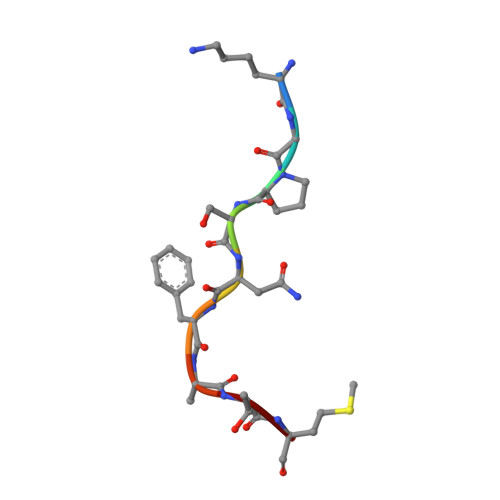
Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
Duru, A.D., Allerbring, E.B., Uchtenhagen, H., Mazumdar, P.A., Badia-Martinez, D., Madhurantakam, C., Sandalova, T., Nygren, P., Achour, A.To be published.