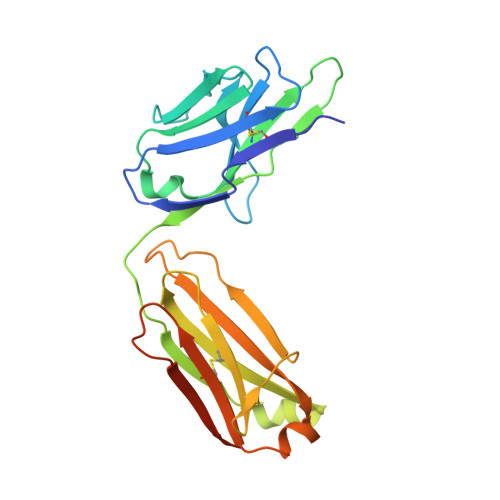
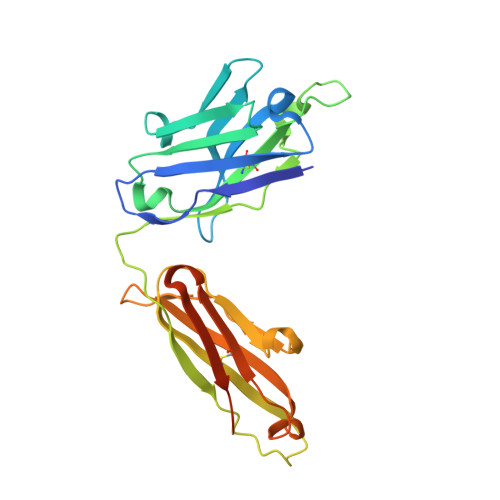
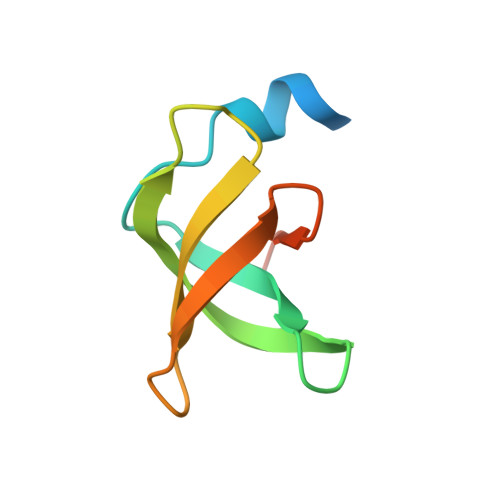
CDR-H3 Diversity Is Not Required for Antigen Recognition by Synthetic Antibodies.
Persson, H., Ye, W., Wernimont, A., Adams, J.J., Koide, A., Koide, S., Lam, R., Sidhu, S.S.(2013) J Mol Biol 425: 803-811
- PubMed: 23219464
- DOI: https://doi.org/10.1016/j.jmb.2012.11.037
- Primary Citation of Related Structures:
3PNW - PubMed Abstract:
A synthetic phage-displayed antibody repertoire was constructed with equivalent chemical diversity in the third complementarity-determining regions of the heavy (CDR-H3) and light (CDR-L3) chains, which contrasts with natural antibodies in which CDR-H3 is much more diverse than CDR-L3 due to the genetic mechanisms that generate antibody encoding genes. Surprisingly, the synthetic repertoire yielded numerous functional antibodies that contained mutated CDR-L3 sequences but a fixed CDR-H3 sequence. Alanine-scanning analysis of antibodies that recognized 10 different antigens but contained a common CDR-H3 loop showed that, in most cases, the fixed CDR-H3 sequence was able to contribute favorably to antigen recognition, but in some cases, the loop was functionally inert. Structural analysis of one such antibody in complex with antigen showed that the inert CDR-H3 loop was nonetheless highly buried at the antibody-antigen interface. Taken together, these results show that CDR-H3 diversity is not necessarily required for the generation of antibodies that recognize diverse protein antigens with high affinity and specificity, and if given the chance, CDR-L3 readily assumes the dominant role for antigen recognition. These results contrast with the commonly accepted view of antigen recognition derived from the analysis of natural antibodies, in which CDR-H3 is presumed to be dominant and CDR-L3 is presumed to play an auxiliary role. Furthermore, the results show that natural antibody function is genetically constrained, and it should be possible to develop more functional synthetic antibody libraries by expanding the diversity of CDR-L3 beyond what is observed in nature.
Organizational Affiliation:
Banting and Best Department of Medical Research and Department of Molecular Genetics, The Donnelly Centre, University of Toronto, 160 College Street, Toronto, Ontario, Canada M5S 3E1.