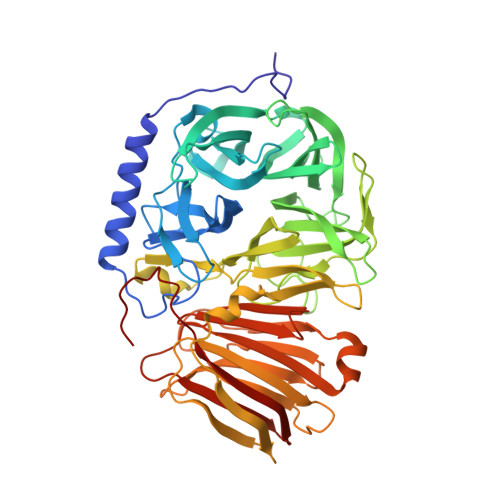
Crystal structures of the apo form of beta-fructofuranosidase from Bifidobacterium longum and its complex with fructose
Bujacz, A., Jedrzejczak-Krzepkowska, M., Bielecki, S., Redzynia, I., Bujacz, G.(2011) FEBS J 278: 1728-1744
- PubMed: 21418142
- DOI: https://doi.org/10.1111/j.1742-4658.2011.08098.x
- Primary Citation of Related Structures:
3PIG, 3PIJ - PubMed Abstract:
We solved the 1.8 Å crystal structure of β-fructofuranosidase from Bifidobacterium longum KN29.1 - a unique enzyme that allows these probiotic bacteria to function in the human digestive system. The sequence of β-fructofuranosidase classifies it as belonging to the glycoside hydrolase family 32 (GH32). GH32 enzymes show a wide range of substrate specificity and different functions in various organisms. All enzymes from this family share a similar fold, containing two domains: an N-terminal five-bladed β-propeller and a C-terminal β-sandwich module. The active site is located in the centre of the β-propeller domain, in the bottom of a 'funnel'. The binding site, -1, responsible for tight fructose binding, is highly conserved among the GH32 enzymes. Bifidobacterium longum KN29.1 β-fructofuranosidase has a 35-residue elongation of the N-terminus containing a five-turn α-helix, which distinguishes it from the other known members of the GH32 family. This new structural element could be one of the functional modifications of the enzyme that allows the bacteria to act in a human digestive system. We also solved the 1.8 Å crystal structure of the β-fructofuranosidase complex with β-D-fructose, a hydrolysis product obtained by soaking apo crystal in raffinose.
Organizational Affiliation:
Institute of Technical Biochemistry, Faculty of Biotechnology and Food Sciences, Technical University of Lodz, Lodz, Poland. anna.bujacz@p.lodz.pl