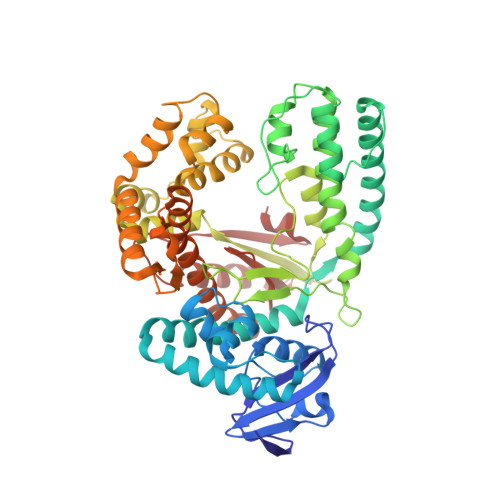
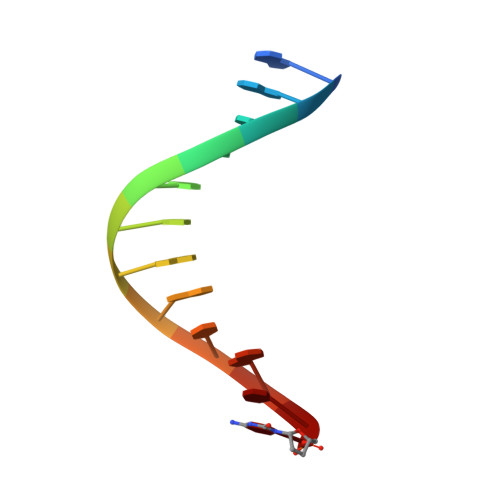
Structural basis for the synthesis of nucleobase modified DNA by Thermus aquaticus DNA polymerase.
Obeid, S., Baccaro, A., Welte, W., Diederichs, K., Marx, A.(2010) Proc Natl Acad Sci U S A 107: 21327-21331
- PubMed: 21123743
- DOI: https://doi.org/10.1073/pnas.1013804107
- Primary Citation of Related Structures:
3OJS, 3OJU - PubMed Abstract:
Numerous 2'-deoxynucleoside triphosphates (dNTPs) that are functionalized with spacious modifications such as dyes and affinity tags like biotin are substrates for DNA polymerases. They are widely employed in many cutting-edge technologies like advanced DNA sequencing approaches, microarrays, and single molecule techniques. Modifications attached to the nucleobase are accepted by many DNA polymerases, and thus, dNTPs bearing nucleobase modifications are predominantly employed. When pyrimidines are used the modifications are almost exclusively at the C5 position to avoid disturbing of Watson-Crick base pairing ability. However, the detailed molecular mechanism by which C5 modifications are processed by a DNA polymerase is poorly understood. Here, we present the first crystal structures of a DNA polymerase from Thermus aquaticus processing two C5 modified substrates that are accepted by the enzyme with different efficiencies. The structures were obtained as ternary complex of the enzyme bound to primer/template duplex with the respective modified dNTP in position poised for catalysis leading to incorporation. Thus, the study provides insights into the incorporation mechanism of the modified nucleotides elucidating how bulky modifications are accepted by the enzyme. The structures show a varied degree of perturbation of the enzyme substrate complexes depending on the nature of the modifications suggesting design principles for future developments of modified substrates for DNA polymerases.
Organizational Affiliation:
Department of Chemistry, Konstanz Research School Chemical Biology, University of Konstanz, Universitätsstrasse 10, D 78457 Konstanz, Germany.