Investigation of the Electron Transport Chain to and the Catalytic Activity of the Diheme Cytochrome c Peroxidase CcpA of Shewanella oneidensis.
Schutz, B., Seidel, J., Sturm, G., Einsle, O., Gescher, J.(2011) Appl Environ Microbiol 77: 6172-6180
- PubMed: 21742904
- DOI: https://doi.org/10.1128/AEM.00606-11
- Primary Citation of Related Structures:
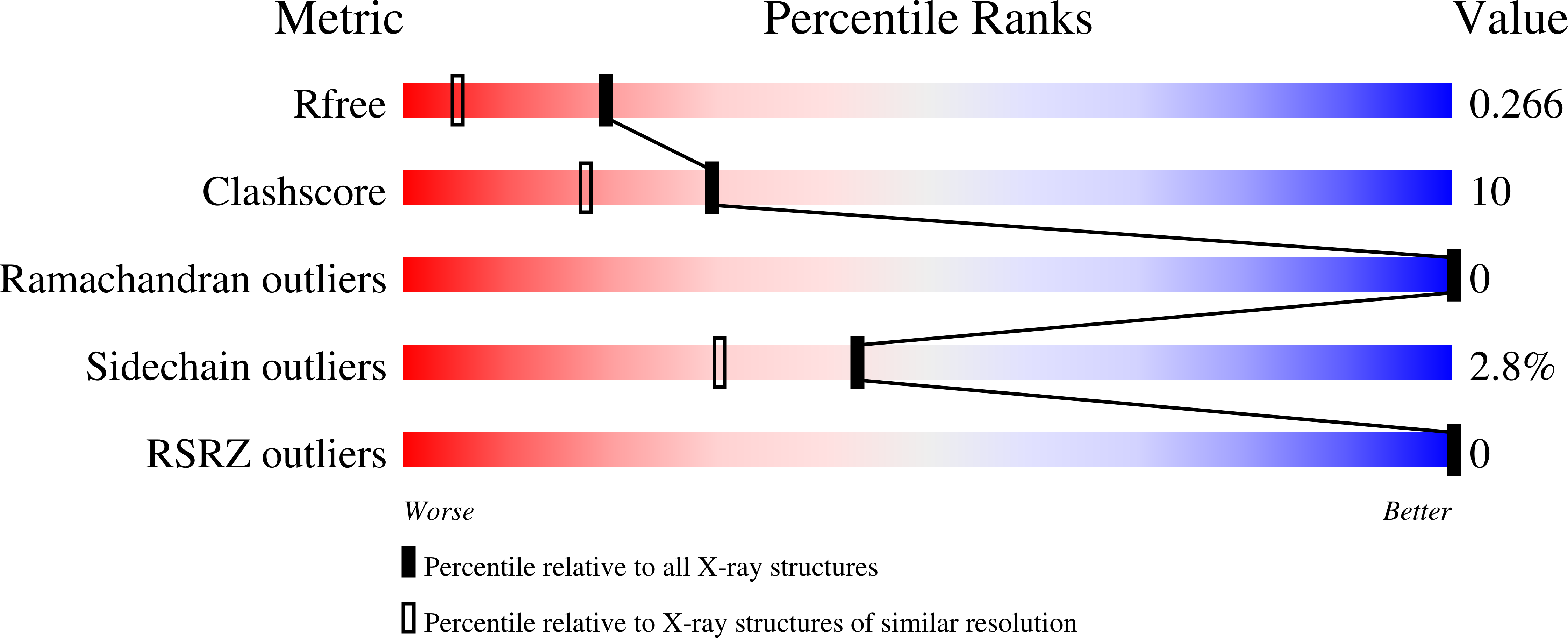
3O5C - PubMed Abstract:
Bacterial diheme c-type cytochrome peroxidases (BCCPs) catalyze the periplasmic reduction of hydrogen peroxide to water. The gammaproteobacterium Shewanella oneidensis produces the peroxidase CcpA under a number of anaerobic conditions, including dissimilatory iron-reducing conditions. We wanted to understand the function of this protein in the organism and its putative connection to the electron transport chain to ferric iron. CcpA was isolated and tested for peroxidase activity, and its structural conformation was analyzed by X-ray crystallography. CcpA exhibited in vitro peroxidase activity and had a structure typical of diheme peroxidases. It was produced in almost equal amounts under anaerobic and microaerophilic conditions. With 50 mM ferric citrate and 50 μM oxygen in the growth medium, CcpA expression results in a strong selective advantage for the cell, which was detected in competitive growth experiments with wild-type and ΔccpA mutant cells that lack the entire ccpA gene due to a markerless deletion. We were unable to reduce CcpA directly with CymA, MtrA, or FccA, which are known key players in the chain of electron transport to ferric iron and fumarate but identified the small monoheme ScyA as a mediator of electron transport between CymA and BCCP. To our knowledge, this is the first detailed description of a complete chain of electron transport to a periplasmic c-type cytochrome peroxidase. This study furthermore reports the possibility of establishing a specific electron transport chain using c-type cytochromes.
Organizational Affiliation:
Institut für Biologie II, Mikrobiologie, Universität Freiburg, Schänzlestr. 1, D-79104 Freiburg, Germany.