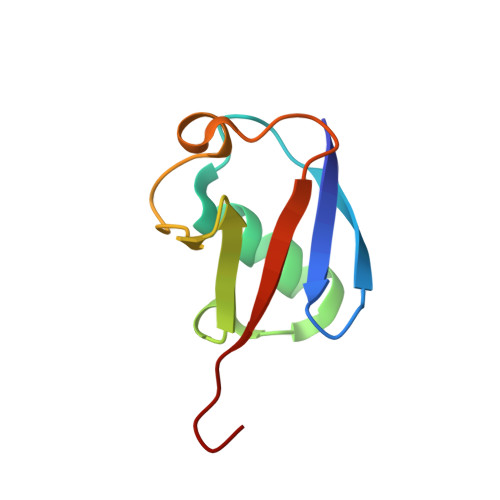
A new crystal form of Lys48-linked diubiquitin.
Trempe, J.F., Brown, N.R., Noble, M.E., Endicott, J.A.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 994-998
- PubMed: 20823512
- DOI: https://doi.org/10.1107/S1744309110027600
- Primary Citation of Related Structures:
3M3J - PubMed Abstract:
Lys48-linked polyubiquitin chains are recognized by the proteasome as a tag for the degradation of the attached substrates. Here, a new crystal form of Lys48-linked diubiquitin (Ub2) was obtained and the crystal structure was refined to 1.6 A resolution. The structure reveals an ordered isopeptide bond in a trans configuration. All three molecules in the asymmetric unit were in the same closed conformation, in which the hydrophobic patches of both the distal and the proximal moieties interact with each other. Despite the different crystallization conditions and different crystal packing, the new crystal structure of Ub2 is similar to the previously published structure of diubiquitin, but differences are observed in the conformation of the flexible isopeptide linkage.
Organizational Affiliation:
Department of Biochemistry, McGill University, 3649 Promenade Sir William Osler, Montreal, Québec H3G 0B1, Canada. jean.trempe@mail.mcgill.ca