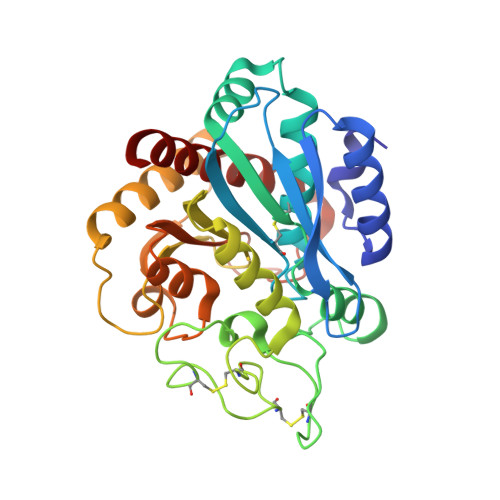
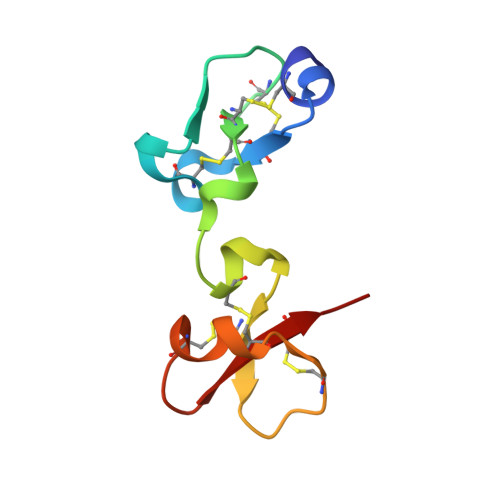
Insights into the molecular inactivation mechanism of human activated thrombin-activatable fibrinolysis inhibitor
Sanglas, L., Arolas, J.L., Valnickova, Z., Aviles, F.X., Enghild, J.J., Gomis-Ruth, F.X.(2010) J Thromb Haemost 8: 1056-1065
- PubMed: 20088943
- DOI: https://doi.org/10.1111/j.1538-7836.2010.03740.x
- Primary Citation of Related Structures:
3LMS - PubMed Abstract:
Thrombin-activatable fibrinolysis inhibitor (TAFI) is a validated target for thrombotic diseases. TAFI is converted in vivo to activated TAFI (TAFIa) by removal of its pro-domain. Whereas TAFI is stable and persists in the circulation, possibly in complex with plasminogen, TAFIa is unstable and poorly soluble, with a half-life of minutes. In order to study the molecular determinants of this instability, we studied the influence of protein inhibitors on human TAFIa. We found that protein inhibitors significantly reduced the instability and insolubility of TAFIa. In addition, we solved the 2.5-A resolution crystal structure of human TAFIa in complex with a potent protein inhibitor, tick-derived carboxypeptidase inhibitor, which gives rise to a stable and soluble TAFIa species. The structure revealed a significant reduction in the flexibility of dynamic segments when compared with the structures of bovine and human TAFI. We also identified two latent hotspots, loop Lbeta2beta3 and segment alpha5-Lalpha5beta7-beta7, where conformational destabilization may begin. These hotspots are also present in TAFI, but the pro-domain may provide sufficient stabilization and solubility to guarantee protein persistence in vivo. When the pro-domain is removed, the free TAFIa moiety becomes unstable, its activity is suppressed, and the molecule becomes insoluble. The present study corroborates the function of protein inhibitors in stabilizing human TAFIa and it provides a rigid and high-resolution mold for the design of small molecule inhibitors of this enzyme, thus paving the way for novel therapy for thrombotic disorders.
Organizational Affiliation:
Departament de Bioquímica i Biologia Molecular, Facultat de Ciències, Institut de Biotecnologia i de Biomedicina, Universitat Autònoma de Barcelona, Bellaterra, Spain.