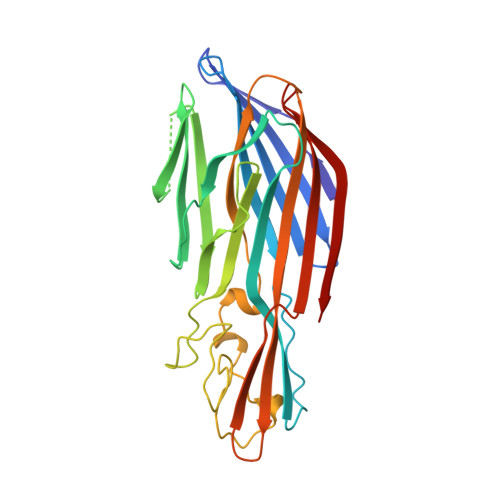
Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel.
Olson, R., Nariya, H., Yokota, K., Kamio, Y., Gouaux, E.(1999) Nat Struct Biol 6: 134-140
- PubMed: 10048924
- DOI: https://doi.org/10.1038/5821
- Primary Citation of Related Structures:
1LKF, 2LKF, 3LKF - PubMed Abstract:
Staphylococcal LukF, LukS, HgammaII, and alpha-hemolysin are self-assembling, channel-forming proteins related in sequence and function. In the alpha-hemolysin heptamer, the channel-forming beta-strands and the amino latch make long excursions from the protomer core. Here we report the crystal structure of the water soluble form of LukF. In the LukF structure the channel-forming region folds into an amphipathic, three-strand beta-sheet and the amino latch forms a beta-strand extending a central beta-sheet. The LukF structure illustrates how a channel-forming toxin masks protein-protein and protein-membrane interfaces prior to cell binding and assembly, and together with the alpha-hemolysin heptamer structure, they define the end points on the pathway of toxin assembly.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, New York 10032, USA.