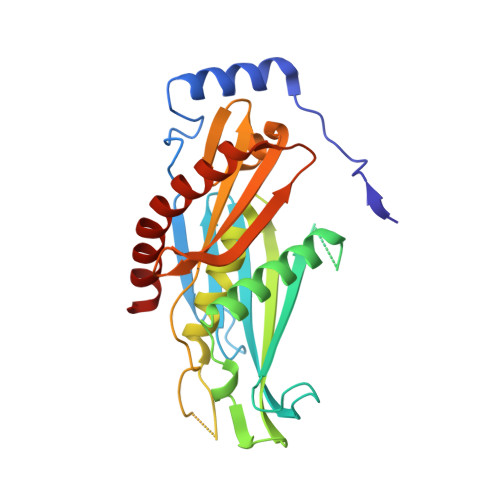
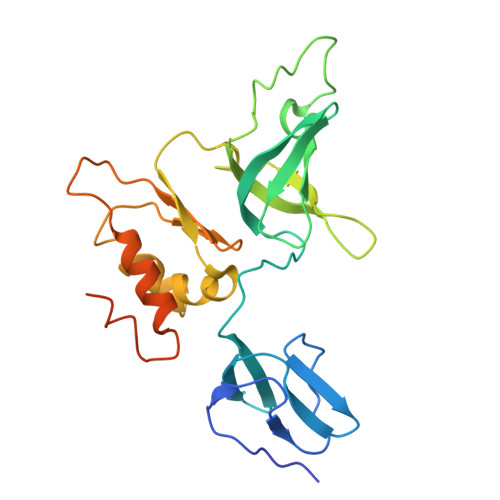
Crystal structure of the S. solfataricus archaeal exosome reveals conformational flexibility in the RNA-binding ring.
Lu, C., Ding, F., Ke, A.(2010) PLoS One 5: e8739-e8739
- PubMed: 20090900
- DOI: https://doi.org/10.1371/journal.pone.0008739
- Primary Citation of Related Structures:
3L7Z - PubMed Abstract:
The exosome complex is an essential RNA 3'-end processing and degradation machinery. In archaeal organisms, the exosome consists of a catalytic ring and an RNA-binding ring, both of which were previously reported to assume three-fold symmetry. Here we report an asymmetric 2.9 A Sulfolobus solfataricus archaeal exosome structure in which the three-fold symmetry is broken due to combined rigid body and thermal motions mainly within the RNA-binding ring. Since increased conformational flexibility was also observed in the RNA-binding ring of the related bacterial PNPase, we speculate that this may reflect an evolutionarily conserved mechanism to accommodate diverse RNA substrates for degradation. This study clearly shows the dynamic structures within the RNA-binding domains, which provides additional insights on mechanism of asymmetric RNA binding and processing.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York, United States of America.