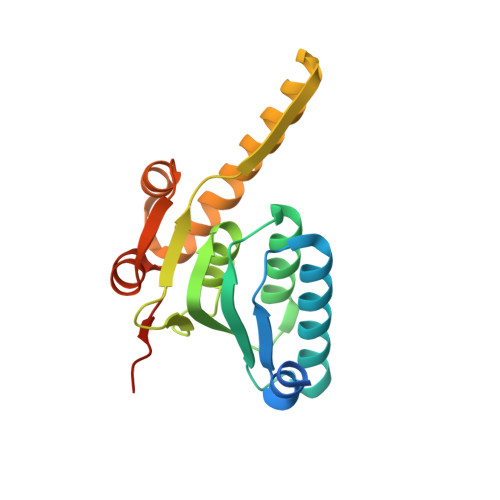
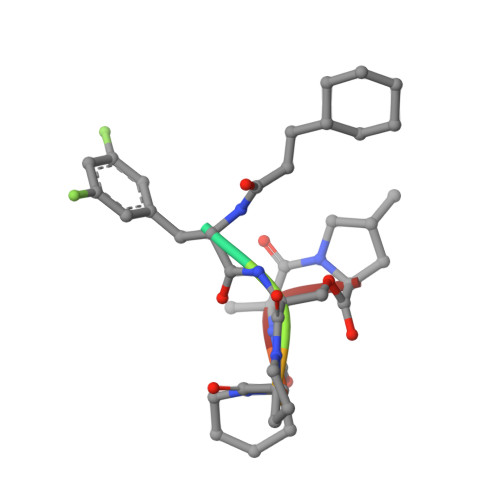
Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism
Lee, B.-G., Park, E.Y., Lee, K.-E., Jeon, H., Sung, K.H., Paulsen, H., Rubsamen-Schaeff, H., Brotz-Oesterhelt, H., Song, H.K.(2010) Nat Struct Mol Biol 17: 471-478
- PubMed: 20305655
- DOI: https://doi.org/10.1038/nsmb.1787
- Primary Citation of Related Structures:
3KTG, 3KTH, 3KTI, 3KTJ, 3KTK - PubMed Abstract:
Clp-family proteins are prototypes for studying the mechanism of ATP-dependent proteases because the proteolytic activity of the ClpP core is tightly regulated by activating Clp-ATPases. Nonetheless, the proteolytic activation mechanism has remained elusive because of the lack of a complex structure. Acyldepsipeptides (ADEPs), a recently discovered class of antibiotics, activate and disregulate ClpP. Here we have elucidated the structural changes underlying the ClpP activation process by ADEPs. We present the structures of Bacillus subtilis ClpP alone and in complex with ADEP1 and ADEP2. The structures show the closed-to-open-gate transition of the ClpP N-terminal segments upon activation as well as conformational changes restricted to the upper portion of ClpP. The direction of the conformational movement and the hydrophobic clustering that stabilizes the closed structure are markedly different from those of other ATP-dependent proteases, providing unprecedented insights into the activation of ClpP.
Organizational Affiliation:
School of Life Sciences and Biotechnology, Korea University, Seoul, Korea.