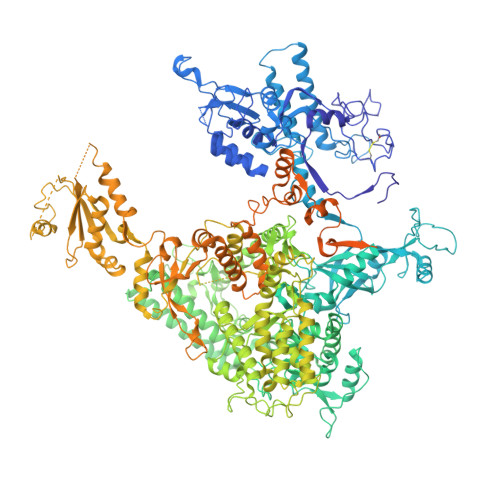
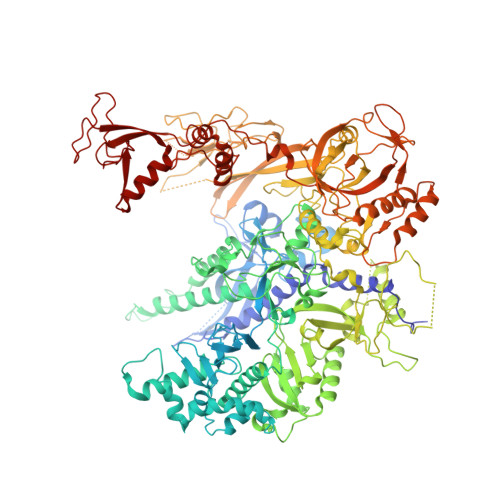
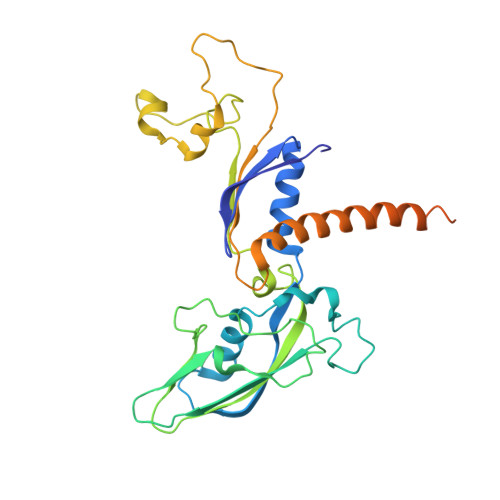
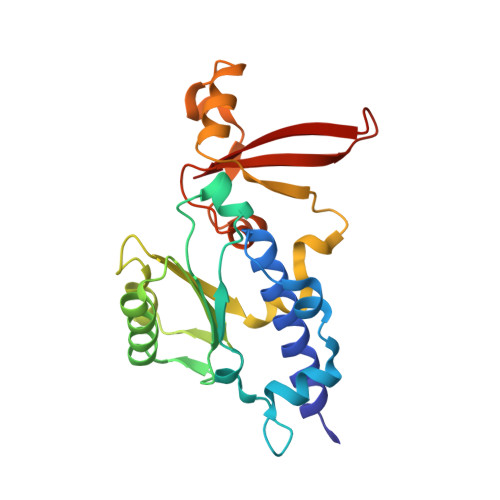
Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism.
Liu, X., Bushnell, D.A., Wang, D., Calero, G., Kornberg, R.D.(2010) Science 327: 206-209
- PubMed: 19965383
- DOI: https://doi.org/10.1126/science.1182015
- Primary Citation of Related Structures:
3K7A - PubMed Abstract:
Previous x-ray crystal structures have given insight into the mechanism of transcription and the role of general transcription factors in the initiation of the process. A structure of an RNA polymerase II-general transcription factor TFIIB complex at 4.5 angstrom resolution revealed the amino-terminal region of TFIIB, including a loop termed the "B finger," reaching into the active center of the polymerase where it may interact with both DNA and RNA, but this structure showed little of the carboxyl-terminal region. A new crystal structure of the same complex at 3.8 angstrom resolution obtained under different solution conditions is complementary with the previous one, revealing the carboxyl-terminal region of TFIIB, located above the polymerase active center cleft, but showing none of the B finger. In the new structure, the linker between the amino- and carboxyl-terminal regions can also be seen, snaking down from above the cleft toward the active center. The two structures, taken together with others previously obtained, dispel long-standing mysteries of the transcription initiation process.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.