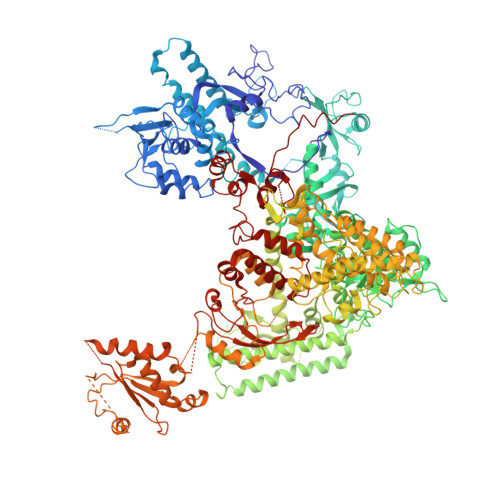
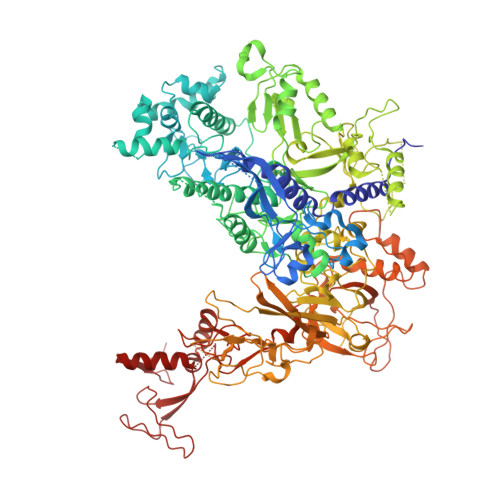
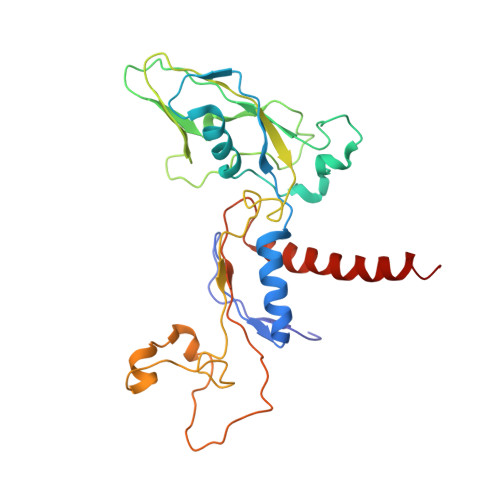
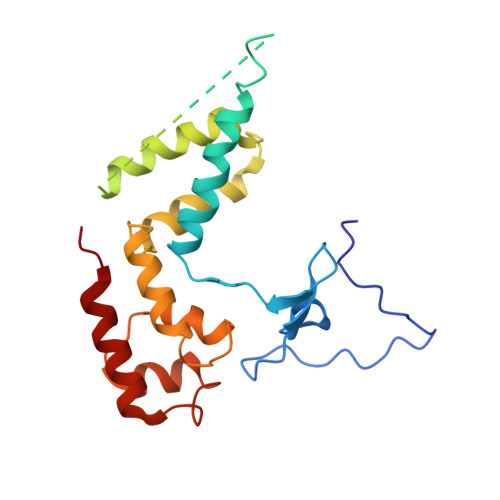
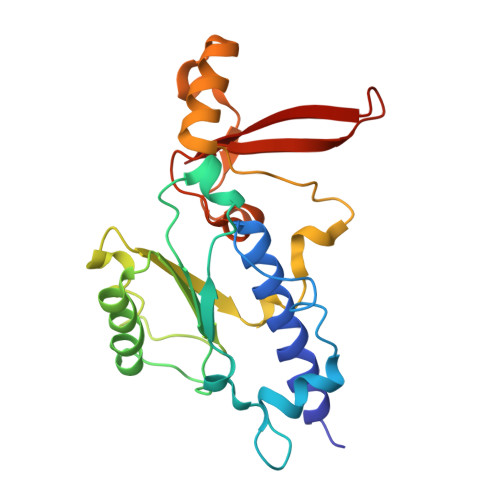
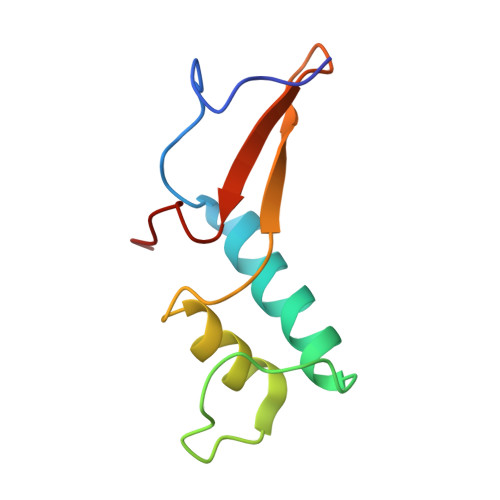
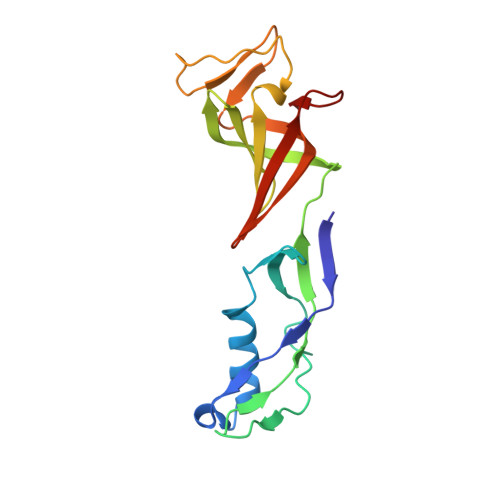
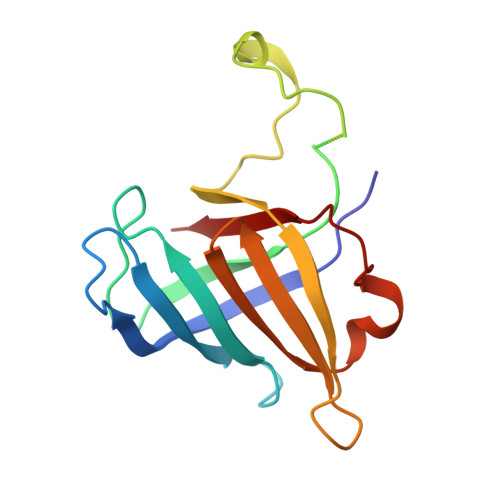
Interaction of the mediator head module with RNA polymerase II.
Cai, G., Chaban, Y.L., Imasaki, T., Kovacs, J.A., Calero, G., Penczek, P.A., Takagi, Y., Asturias, F.J.(2012) Structure 20: 899-910
- PubMed: 22579255
- DOI: https://doi.org/10.1016/j.str.2012.02.023
- Primary Citation of Related Structures:
3J1N, 3J1O - PubMed Abstract:
Mediator, a large (21 polypeptides, MW ∼1 MDa) complex conserved throughout eukaryotes, plays an essential role in control of gene expression by conveying regulatory signals that influence the activity of the preinitiation complex. However, the precise mode of interaction between Mediator and RNA polymerase II (RNAPII), and the mechanism of regulation by Mediator remain elusive. We used cryo-electron microscopy and reconstituted in vitro transcription assays to characterize a transcriptionally-active complex including the Mediator Head module and components of a minimum preinitiation complex (RNAPII, TFIIF, TFIIB, TBP, and promoter DNA). Our results reveal how the Head interacts with RNAPII, affecting its conformation and function.
Organizational Affiliation:
Department of Cell Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.