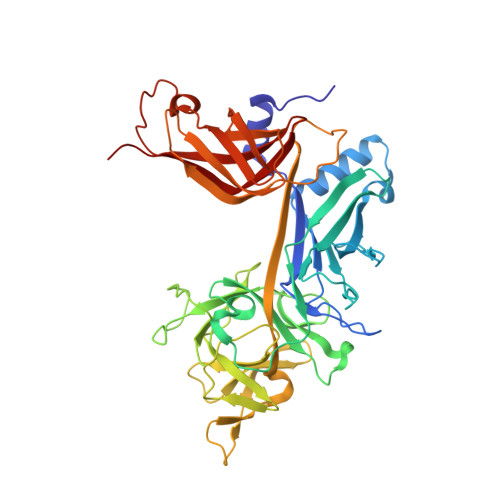
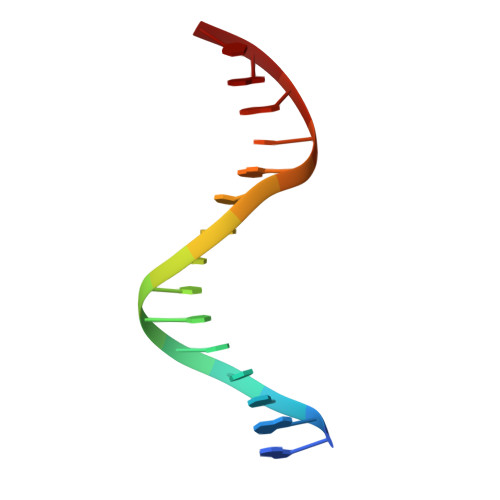
Thermodynamic and structural insights into CSL-DNA complexes.
Friedmann, D.R., Kovall, R.A.(2010) Protein Sci 19: 34-46
- PubMed: 19866488
- DOI: https://doi.org/10.1002/pro.280
- Primary Citation of Related Structures:
3IAG - PubMed Abstract:
The Notch pathway is an intercellular signaling mechanism that plays important roles in cell fates decisions throughout the developing and adult organism. Extracellular complexation of Notch receptors with ligands ultimately results in changes in gene expression, which is regulated by the nuclear effector of the pathway, CSL (C-promoter binding factor 1 (CBF-1), suppressor of hairless (Su(H)), lin-12 and glp-1 (Lag-1)). CSL is a DNA binding protein that is involved in both repression and activation of transcription from genes that are responsive to Notch signaling. One well-characterized Notch target gene is hairy and enhancer of split-1 (HES-1), which is regulated by a promoter element consisting of two CSL binding sites oriented in a head-to-head arrangement. Although previous studies have identified in vivo and consensus binding sites for CSL, and crystal structures of these complexes have been determined, to date, a quantitative description of the energetics that underlie CSL-DNA binding is unknown. Here, we provide a thermodynamic and structural analysis of the interaction between CSL and the two individual sites that comprise the HES-1 promoter element. Our comprehensive studies that analyze binding as a function of temperature, salt, and pH reveal moderate, but distinct, differences in the affinities of CSL for the two HES-1 binding sites. Similarly, our structural results indicate that overall CSL binds both DNA sites in a similar manner; however, minor changes are observed in both the conformation of CSL and DNA. Taken together, our results provide a quantitative and biophysical basis for understanding how CSL interacts with DNA sites in vivo.
Organizational Affiliation:
Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati College of Medicine, Cincinnati, Ohio 45267-0524, USA.