Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance.
Ghosh, A.K., Kulkarni, S., Anderson, D.D., Hong, L., Baldridge, A., Wang, Y.F., Chumanevich, A.A., Kovalevsky, A.Y., Tojo, Y., Amano, M., Koh, Y., Tang, J., Weber, I.T., Mitsuya, H.(2009) J Med Chem 52: 7689-7705
- PubMed: 19746963
- DOI: https://doi.org/10.1021/jm900695w
- Primary Citation of Related Structures:
3I6O, 3I7E - PubMed Abstract:
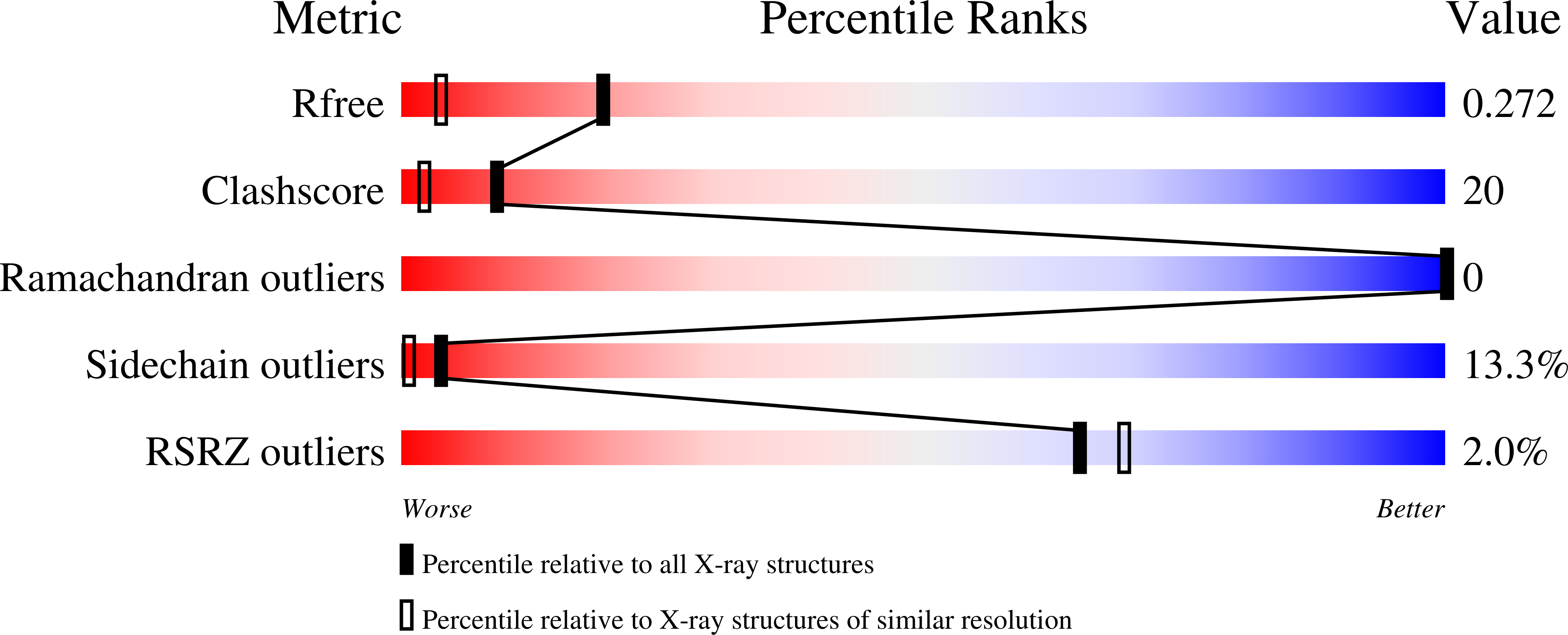
The structure-based design, synthesis, and biological evaluation of a series of nonpeptidic macrocyclic HIV protease inhibitors are described. The inhibitors are designed to effectively fill in the hydrophobic pocket in the S1'-S2' subsites and retain all major hydrogen bonding interactions with the protein backbone similar to darunavir (1) or inhibitor 2. The ring size, the effect of methyl substitution, and unsaturation within the macrocyclic ring structure were assessed. In general, cyclic inhibitors were significantly more potent than their acyclic homologues, saturated rings were less active than their unsaturated analogues and a preference for 10- and 13-membered macrocylic rings was revealed. The addition of methyl substituents resulted in a reduction of potency. Both inhibitors 14b and 14c exhibited marked enzyme inhibitory and antiviral activity, and they exerted potent activity against multidrug-resistant HIV-1 variants. Protein-ligand X-ray structures of inhibitors 2 and 14c provided critical molecular insights into the ligand-binding site interactions.
Organizational Affiliation:
Department of Chemistry and Medicinal Chemistry, Purdue University, West Lafayette, Indiana 47907, USA. akghosh@purdue.edu