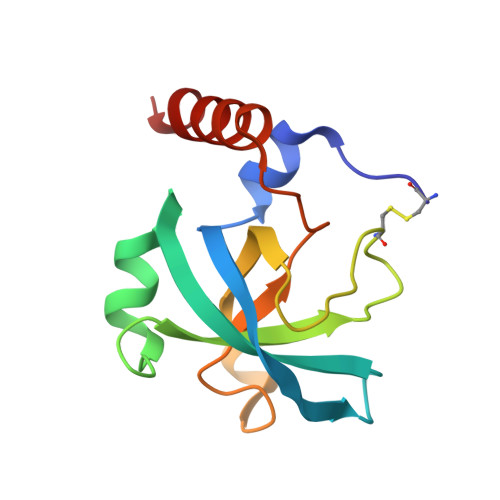
An interdomain disulfide bridge links the NtA and first FS domain in agrin.
McFarlane, A.A., Stetefeld, J.(2009) Protein Sci 18: 2421-2428
- PubMed: 19845005
- DOI: https://doi.org/10.1002/pro.276
- Primary Citation of Related Structures:
3I70 - PubMed Abstract:
Agrin is a multidomain heparan sulfate proteoglycan involved in postsynaptic differentiation at the neuromuscular junction. Binding of agrin to synaptic basal lamina is mediated by the N-terminal agrin (NtA) domain. The NtA domain of agrin is followed by a tandem of nine follistatin-like (FS) domains forming a rod-like spacer to the laminin G-like domains of the molecule. Here we report that the most C-terminal cysteine residue of NtA (Cys123) forms an interdomain disulfide bond with the FOLN subdomain of the FS module. Remarkably, this single cysteine is flanked by Leu117 and Val124, which are two essential beta-branched amino acids forming the heterocomplex of NtA with the gamma 1 chain of laminin. Moreover, we show that this covalent linkage compensates for the seven amino acid residue splice insert at the very C-terminal helix H3 and causes a rigid interface between NtA and FS independent of the alternative mRNA splice event. These results suggest that the interdomain disulfide bond between the NtA and the first FS domain might be important for the proper folding of agrin.
Organizational Affiliation:
Department of Chemistry, University of Manitoba, Winnipeg, Manitoba R3T 2N2, Canada.