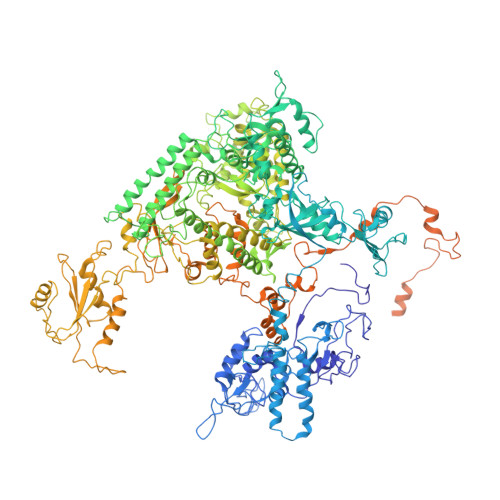
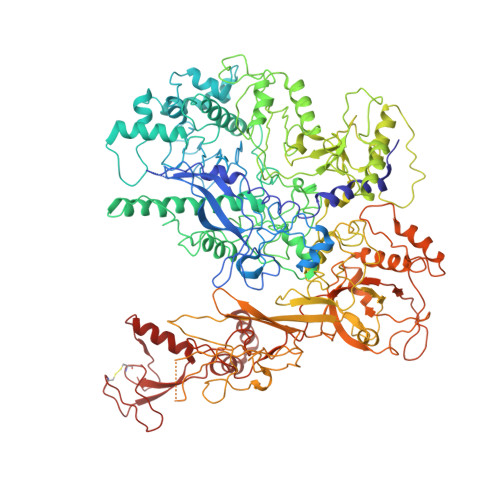
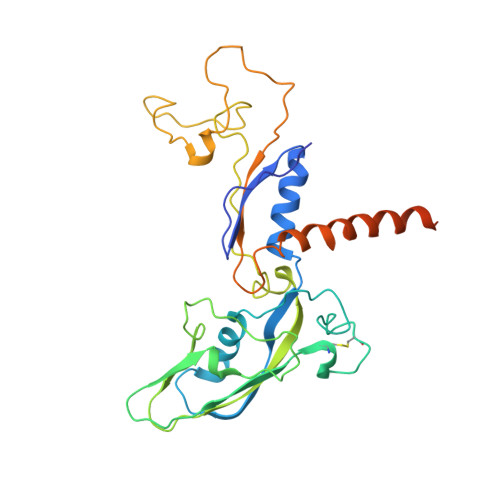
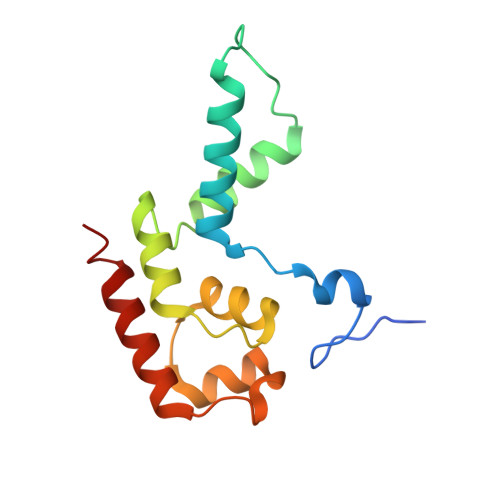
Schizosacharomyces pombe RNA polymerase II at 3.6-A resolution.
Spahr, H., Calero, G., Bushnell, D.A., Kornberg, R.D.(2009) Proc Natl Acad Sci U S A 106: 9185-9190
- PubMed: 19458260
- DOI: https://doi.org/10.1073/pnas.0903361106
- Primary Citation of Related Structures:
3H0G - PubMed Abstract:
The second structure of a eukaryotic RNA polymerase II so far determined, that of the enzyme from the fission yeast Schizosaccharomyces pombe, is reported here. Comparison with the previous structure of the enzyme from the budding yeast Saccharomyces cerevisiae reveals differences in regions implicated in start site selection and transcription factor interaction. These aspects of the transcription mechanism differ between S. pombe and S. cerevisiae, but are conserved between S. pombe and humans. Amino acid changes apparently responsible for the structural differences are also conserved between S. pombe and humans, suggesting that the S. pombe structure may be a good surrogate for that of the human enzyme.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.