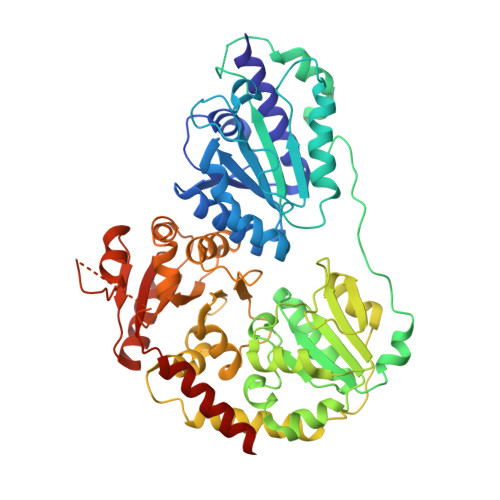
Structural insights of the MenD from Escherichia coli reveal ThDP affinity.
Priyadarshi, A., Saleem, Y., Nam, K.H., Kim, K.S., Park, S.Y., Kim, E.E., Hwang, K.Y.(2009) Biochem Biophys Res Commun 380: 797-801
- PubMed: 19338755
- DOI: https://doi.org/10.1016/j.bbrc.2009.01.168
- Primary Citation of Related Structures:
3FLM - PubMed Abstract:
MenD (2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate) synthase belongs to the superfamily of thiamin diphosphate-dependent decarboxylases, which converts isochorismate and 2-oxoglutarate to SHCHC, pyruvate, and carbon dioxide. Here, we report the first crystal structure of apo-MenD from Escherichia coli determined in tetragonal crystal form. The subunit displays the typical three-domain structure observed for ThDP-dependent enzymes. Analytical gel filtration shows that EcMenD behaves as a dimer as well as a tetramer. Circular dichroism and isothermal calorimetry results confirm EcMenD dependency on ThDP, which concomitantly helps to stabilize with better configuration.
Organizational Affiliation:
Biomedical Research Center, Life Science Division, Korea Institute of Science and Technology, Seongbuk-gu, Seoul 136-791, Republic of Korea.