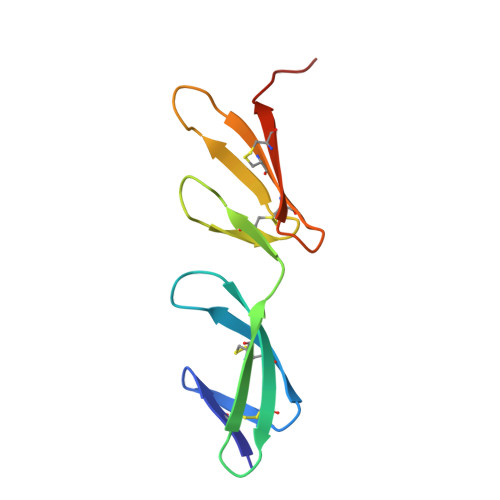
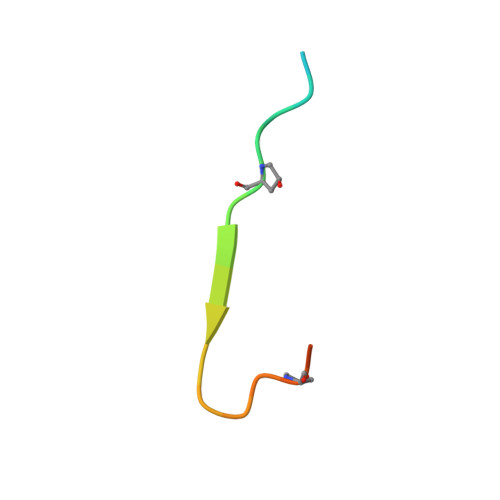
Identification and structural analysis of type I collagen sites in complex with fibronectin fragments.
Erat, M.C., Slatter, D.A., Lowe, E.D., Millard, C.J., Farndale, R.W., Campbell, I.D., Vakonakis, I.(2009) Proc Natl Acad Sci U S A 106: 4195-4200
- PubMed: 19251642
- DOI: https://doi.org/10.1073/pnas.0812516106
- Primary Citation of Related Structures:
3EJH - PubMed Abstract:
Collagen and fibronectin are major components of vertebrate extracellular matrices. Their association and distribution control the development and properties of diverse tissues, but thus far no structural information has been available for the complex formed. Here, we report binding of a peptide, derived from the alpha(1) chain of type I collagen, to the gelatin-binding domain of human fibronectin and present the crystal structure of this peptide in complex with the (8-9)FnI domain pair. Both gelatin-binding domain subfragments, (6)FnI(1-2)FnII(7)FnI and (8-9)FnI, bind the same specific sequence on D-period 4 of collagen I alpha(1), adjacent to the MMP-1 cleavage site. (8-9)FnI also binds the equivalent sequence of the alpha(2) chain. The collagen peptide adopts an antiparallel beta-strand conformation, similar to structures of proteins from pathogenic bacteria bound to FnI domains. Analysis of the type I collagen sequence suggests multiple putative fibronectin-binding sites compatible with our structural model. We demonstrate, by kinetic unfolding experiments, that the triple-helical collagen state is destabilized by (8-9)FnI. This finding suggests a role for fibronectin in collagen proteolysis and tissue remodeling.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, Oxford OX1 3QU, United Kingdom.