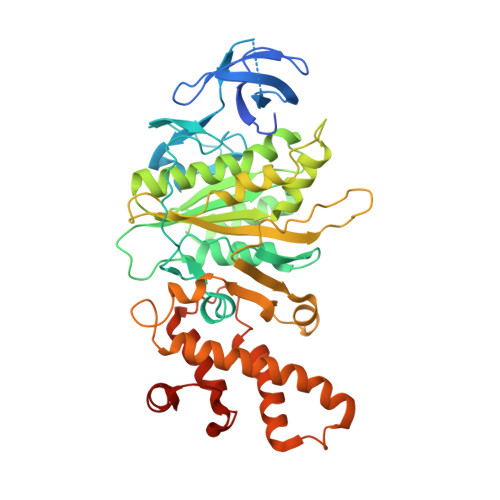
A second transient position of ATP on its trail to the nucleotide-binding site of subunit B of the motor protein A(1)A(O) ATP synthase
Manimekalai, M.S.S., Kumar, A., Balakrishna, A.M., Gruber, G.(2009) J Struct Biol 166: 38-45
- PubMed: 19138746
- DOI: https://doi.org/10.1016/j.jsb.2008.12.004
- Primary Citation of Related Structures:
3EIU - PubMed Abstract:
The adenosine triphosphate (ATP) entrance into the nucleotide-binding subunits of ATP synthases is a puzzle. In the previously determined structure of subunit B mutant R416W of the Methanosarcina mazei Gö1 A-ATP synthase one ATP could be trapped at a transition position, close to the phosphate-binding loop. Using defined parameters for co-crystallization of an ATP-bound B-subunit, a unique transition position of ATP could be found in the crystallographic structure of this complex, solved at 3.4 A resolution. The nucleotide is found near the helix-turn-helix motif in the C-terminal domain of the protein; the location occupied by the gamma-subunit to interact with the empty beta-subunit in the thermoalkaliphilic Bacillus sp. TA2.A1 of the related F-ATP synthase. When compared with the determined structure of the ATP-transition position, close to the P-loop, and the nucleotide-free form of subunit B, the C-terminal domain of the B mutant is rotated by around 6 degrees, implicating an ATP moving pathway. We propose that, in the nucleotide empty state the central stalk subunit D is in close contact with subunit B and when the ATP molecule enters, D moves slightly, paving way for it to interact with the subunit B, which makes the C-terminal domain rotate by 6 degrees.
Organizational Affiliation:
Nanyang Technological University, Division of Structural & Computational Biology, School of Biological Sciences, 60 Nanyang Drive, Singapore 637551, Republic of Singapore.