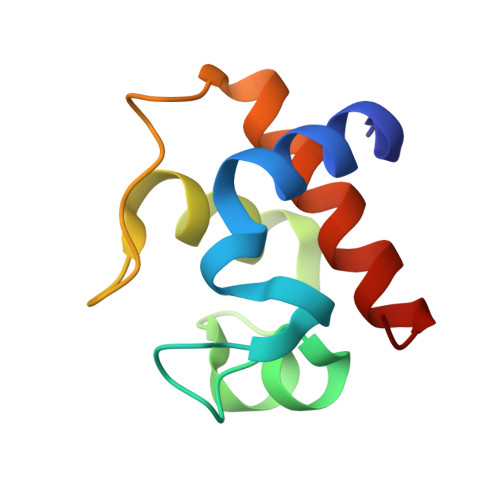
Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Akazaki, H., Kawai, F., Hosokawa, M., Hama, T., Chida, H., Hirano, T., Lim, B.K., Sakurai, N., Hakamata, W., Park, S.Y., Nishio, T., Oku, T.(2009) Biosci Biotechnol Biochem 73: 189-191
- PubMed: 19129656
- DOI: https://doi.org/10.1271/bbb.80472
- Primary Citation of Related Structures:
3DMI - PubMed Abstract:
We determined for the first time the crystal structure of diatom cytochrome c(6) from Phaeodactylum tricornutum at 1.5 A resolution. The overall structure of the protein was classified as a class I c-type cytochrome. The physicochemical properties of the protein were examined by denaturation with guanidine hydrochloride and urea, and compared with those of other algal cytochrome c(6).
Organizational Affiliation:
Bio-Organic Chemistry Laboratory, Graduate School of Bioresource Sciences, Nihon University.