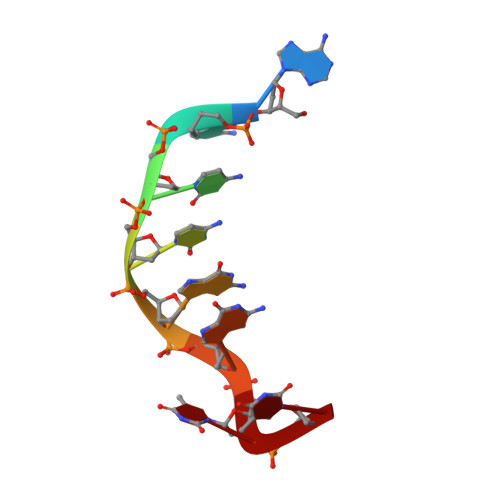
Crystal structure of trioxacarcin A covalently bound to DNA
Pfoh, R., Laatsch, H., Sheldrick, G.M.(2008) Nucleic Acids Res 36: 3508-3514
- PubMed: 18453630
- DOI: https://doi.org/10.1093/nar/gkn245
- Primary Citation of Related Structures:
3C2J - PubMed Abstract:
We report a crystal structure that shows an antibiotic that extracts a nucleobase from a DNA molecule 'caught in the act' after forming a covalent bond but before departing with the base. The structure of trioxacarcin A covalently bound to double-stranded d(AACCGGTT) was determined to 1.78 A resolution by MAD phasing employing brominated oligonucleotides. The DNA-drug complex has a unique structure that combines alkylation (at the N7 position of a guanine), intercalation (on the 3'-side of the alkylated guanine), and base flip-out. An antibiotic-induced flipping-out of a single, nonterminal nucleobase from a DNA duplex was observed for the first time in a crystal structure.
Organizational Affiliation:
Lehrstuhl für Strukturchemie, Georg-August-Universität, Tammannstr. 4, 37077 Göttingen, Germany.