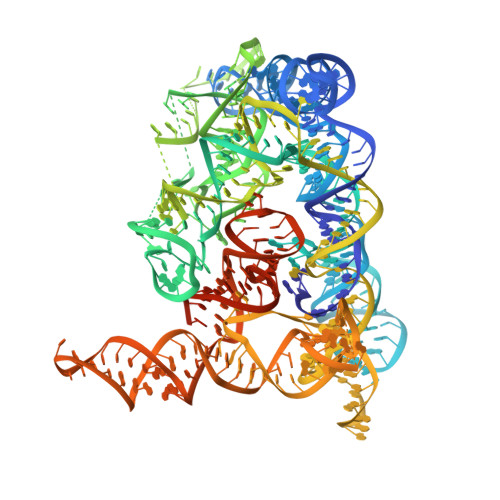
Crystal structure of a self-spliced group II intron
Toor, N., Keating, K.S., Taylor, S.D., Pyle, A.M.(2008) Science 320: 77-82
- PubMed: 18388288
- DOI: https://doi.org/10.1126/science.1153803
- Primary Citation of Related Structures:
3BWP - PubMed Abstract:
Group II introns are self-splicing ribozymes that catalyze their own excision from precursor transcripts and insertion into new genetic locations. Here we report the crystal structure of an intact, self-spliced group II intron from Oceanobacillus iheyensis at 3.1 angstrom resolution. An extensive network of tertiary interactions facilitates the ordered packing of intron subdomains around a ribozyme core that includes catalytic domain V. The bulge of domain V adopts an unusual helical structure that is located adjacent to a major groove triple helix (catalytic triplex). The bulge and catalytic triplex jointly coordinate two divalent metal ions in a configuration that is consistent with a two-metal ion mechanism for catalysis. Structural and functional analogies support the hypothesis that group II introns and the spliceosome share a common ancestor.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, 266 Whitney Avenue, Bass Building, New Haven, CT 06511, USA. navtej.toor@yale.edu