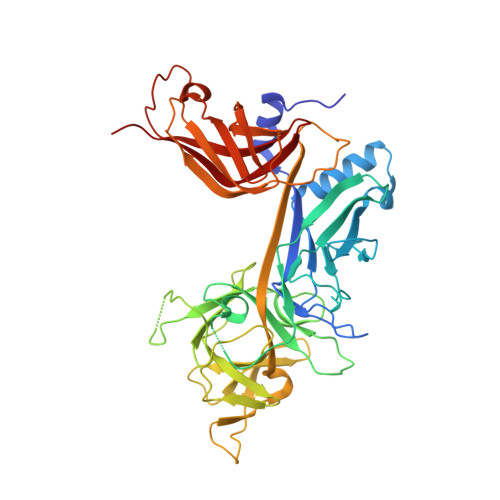
RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
Friedmann, D.R., Wilson, J.J., Kovall, R.A.(2008) J Biol Chem 283: 14781-14791
- PubMed: 18381292
- DOI: https://doi.org/10.1074/jbc.M709501200
- Primary Citation of Related Structures:
3BRD, 3BRF, 3BRG - PubMed Abstract:
The Notch pathway is a conserved cell-to-cell signaling mechanism, in which extracellular signals are transduced into transcriptional outputs through the nuclear effector CSL. CSL is converted from a repressor to an activator through the formation of the CSL-NotchIC-Mastermind ternary complex. The RAM (RBP-J associated molecule) domain of NotchIC avidly interacts with CSL; however, its role in assembly of the CSL-NotchIC-Mastermind ternary complex is not understood. Here we provide a comprehensive thermodynamic, structural, and biochemical analysis of the RAM-CSL interaction for components from both mouse and worm. Our binding data show that RAM and CSL form a high affinity complex in the presence or absence of DNA. Our structural studies reveal a striking distal conformational change in CSL upon RAM binding, which creates a docking site for Mastermind to bind to the complex. Finally, we show that the addition of a RAM peptide in trans facilitates formation of the CSL-NotchIC-Mastermind ternary complex in vitro.
Organizational Affiliation:
Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati College of Medicine, Cincinnati, OH 45267-0524, USA.