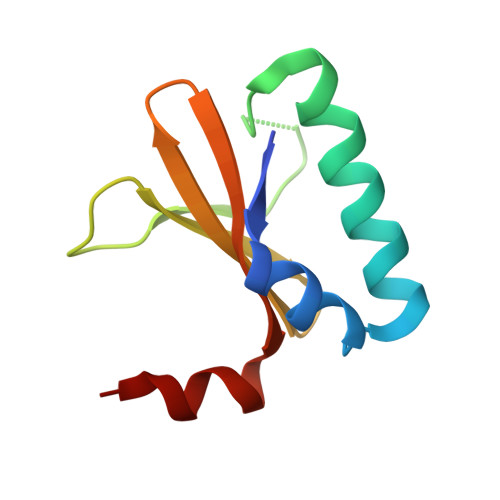
Crystal structure of the antitoxin-toxin protein complex RelB-RelE from Methanococcus jannaschii
Francuski, D., Saenger, W.(2009) J Mol Biol 393: 898-908
- PubMed: 19712680
- DOI: https://doi.org/10.1016/j.jmb.2009.08.048
- Primary Citation of Related Structures:
3BPQ - PubMed Abstract:
Here we present the crystal structure of the Methanococcus jannaschii RelE-RelB (RelBE) toxin-antitoxin (TA) protein complex determined by the MIRAS (multiple isomorphous replacement with anomalous signal) method. The genes encoding this TA system are located in the chromosome of this archaeon and involved in stress response. RelE acts as an endoribonuclease that cleaves mRNA on the ribosome, and we compare the RelBE complex to the known structures of other TA systems belonging to this group and to endoribonucleases. M. jannaschii RelBE forms a heterotetramer with the antitoxin in the centre of the complex, a configuration that differs vastly from the heterotetramer structure of the previously published RelBE from another archaeon, Pyrococcus horikoshii. The long N-terminal alpha-helix of the tightly bound M. jannaschii antitoxin RelB covers the presumed active site of the toxin RelE that is formed by a central beta-sheet, a loop on one side and a C-terminal alpha-helix on the other side. The active site of the M. jannaschii toxin RelE harbours positive charges that are thought to neutralize the negative charges of the substrate mRNA, including Arg62 that was changed to Ser62 by the Escherichia coli expression system, thereby leading to inactive toxin RelE. Comparative studies suggest that Asp43 and His79 are also involved in the activity of the toxin.
Organizational Affiliation:
Kristallographie, Institut für Chemie und Biochemie, Freie Universität Berlin, Takustrasse 6, Berlin 14195, Germany.