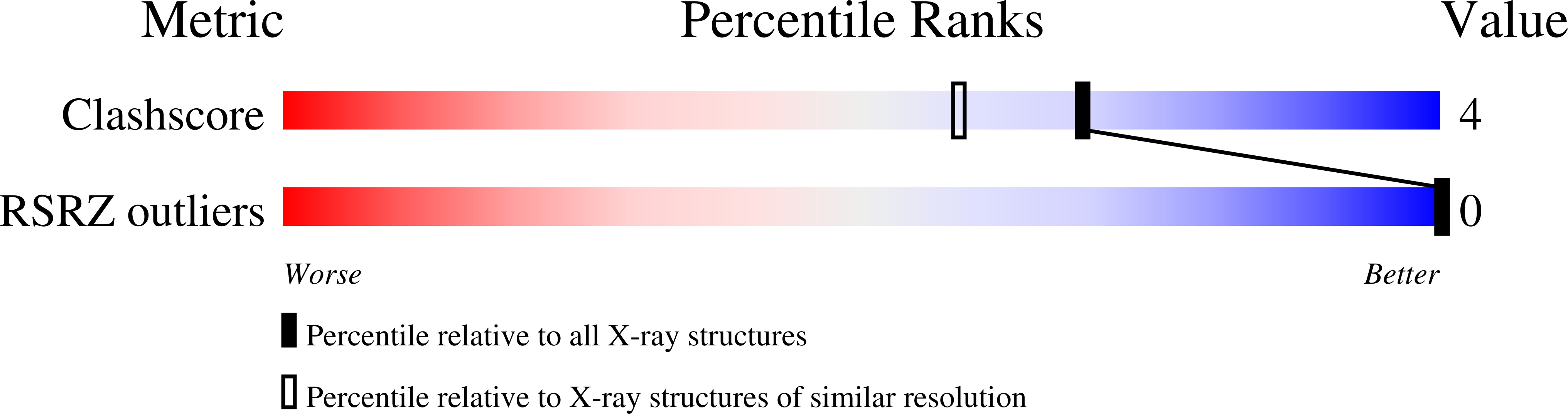Visualisation of extensive water ribbons and networks in a DNA minor-groove drug complex.
Guerri, A., Simpson, I.J., Neidle, S.(1998) Nucleic Acids Res 26: 2873-2878
- PubMed: 9611230
- DOI: https://doi.org/10.1093/nar/26.12.2873
- Primary Citation of Related Structures:
360D - PubMed Abstract:
The crystal structure is reported of a complex between an ethyl derivative of the minor-groove drug furamidine and the dodecanucleotide duplex d(CGCGAATTCGCG)2, which has been refined to 1.85 A resolution and an R factor of 16.6% for data collected at -173 degreesC. An exceptionally large number (220) of water molecules have been located. The majority of these occur in the first coordination shell of solvation. There are extensive networks of connected waters, both in the major and minor grooves. In particular, there are 21 water molecules associated with the minor-groove drug, via hydrogen bonds from the four charged nitrogen atoms. One cluster of four waters is situated in the groove itself; the majority are on the outer edge of the groove, and serve to bridge between the outward-directed drug nitrogen atoms and backbone phosphate oxygen atoms. These bridges are both intra- and inter-strand, with the net effect that the outer edge of the drug molecule is covered by ribbons of water molecules.
Organizational Affiliation:
The CRC Biomolecular Structure Unit, The Institute of Cancer Research, Sutton, Surrey SM2 5NG, UK.