Nonspecific base recognition mediated by water bridges and hydrophobic stacking in ribonuclease I from Escherichia coli
Rodriguez, S.M., Panjikar, S., Van Belle, K., Wyns, L., Messens, J., Loris, R.(2008) Protein Sci 17: 681-690
- PubMed: 18305191
- DOI: https://doi.org/10.1110/ps.073420708
- Primary Citation of Related Structures:
2Z70 - PubMed Abstract:
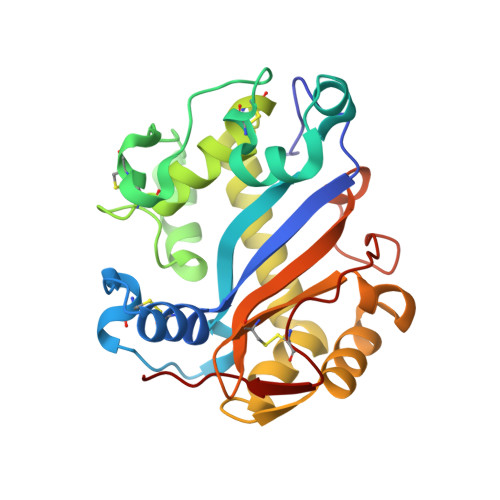
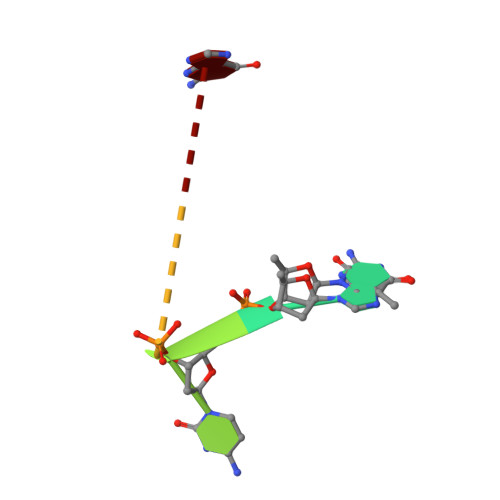
The crystal structure of Escherichia coli ribonuclease I (EcRNase I) reveals an RNase T2-type fold consisting of a conserved core of six beta-strands and three alpha-helices. The overall architecture of the catalytic residues is very similar to the plant and fungal RNase T2 family members, but the perimeter surrounding the active site is characterized by structural elements specific for E. coli. In the structure of EcRNase I in complex with a substrate-mimicking decadeoxynucleotide d(CGCGATCGCG), we observe a cytosine bound in the B2 base binding site and mixed binding of thymine and guanine in the B1 base binding site. The active site residues His55, His133, and Glu129 interact with the phosphodiester linkage only through a set of water molecules. Residues forming the B2 base recognition site are well conserved among bacterial homologs and may generate limited base specificity. On the other hand, the B1 binding cleft acquires true base aspecificity by combining hydrophobic van der Waals contacts at its sides with a water-mediated hydrogen-bonding network at the bottom. This B1 base recognition site is highly variable among bacterial sequences and the observed interactions are unique to EcRNaseI and a few close relatives.
Organizational Affiliation:
Laboratorium voor Ultrastructuur, Vrije Universiteit Brussels, Pleinlaan 2, B-1050 Brussels, Belgium.