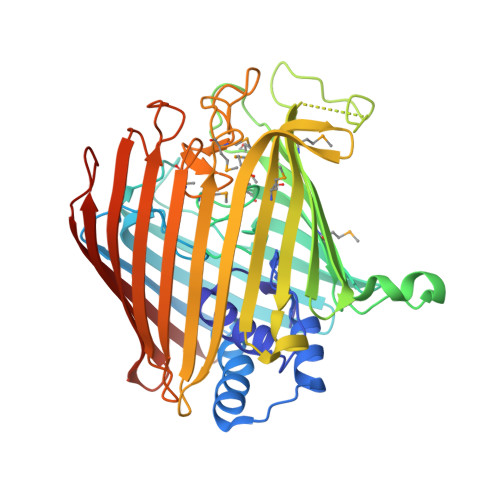
Wzi is an Outer Membrane Lectin that Underpins Group 1 Capsule Assembly in Escherichia Coli.
Bushell, S.R., Mainprize, I.L., Wear, M.A., Lou, H., Whitfield, C., Naismith, J.H.(2013) Structure 21: 844
- PubMed: 23623732
- DOI: https://doi.org/10.1016/j.str.2013.03.010
- Primary Citation of Related Structures:
2YNK - PubMed Abstract:
Many pathogenic bacteria encase themselves in a polysaccharide capsule that provides a barrier to the physical and immunological challenges of the host. The mechanism by which the capsule assembles around the bacterial cell is unknown. Wzi, an integral outer-membrane protein from Escherichia coli, has been implicated in the formation of group 1 capsules. The 2.6 Å resolution structure of Wzi reveals an 18-stranded β-barrel fold with a novel arrangement of long extracellular loops that blocks the extracellular entrance and a helical bundle that plugs the periplasmic end. Mutagenesis shows that specific extracellular loops are required for in vivo capsule assembly. The data show that Wzi binds the K30 carbohydrate polymer and, crucially, that mutants functionally deficient in vivo show no binding to K30 polymer in vitro. We conclude that Wzi is a novel outer-membrane lectin that assists in the formation of the bacterial capsule via direct interaction with capsular polysaccharides.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St Andrews, St Andrews KY16 9ST, UK.