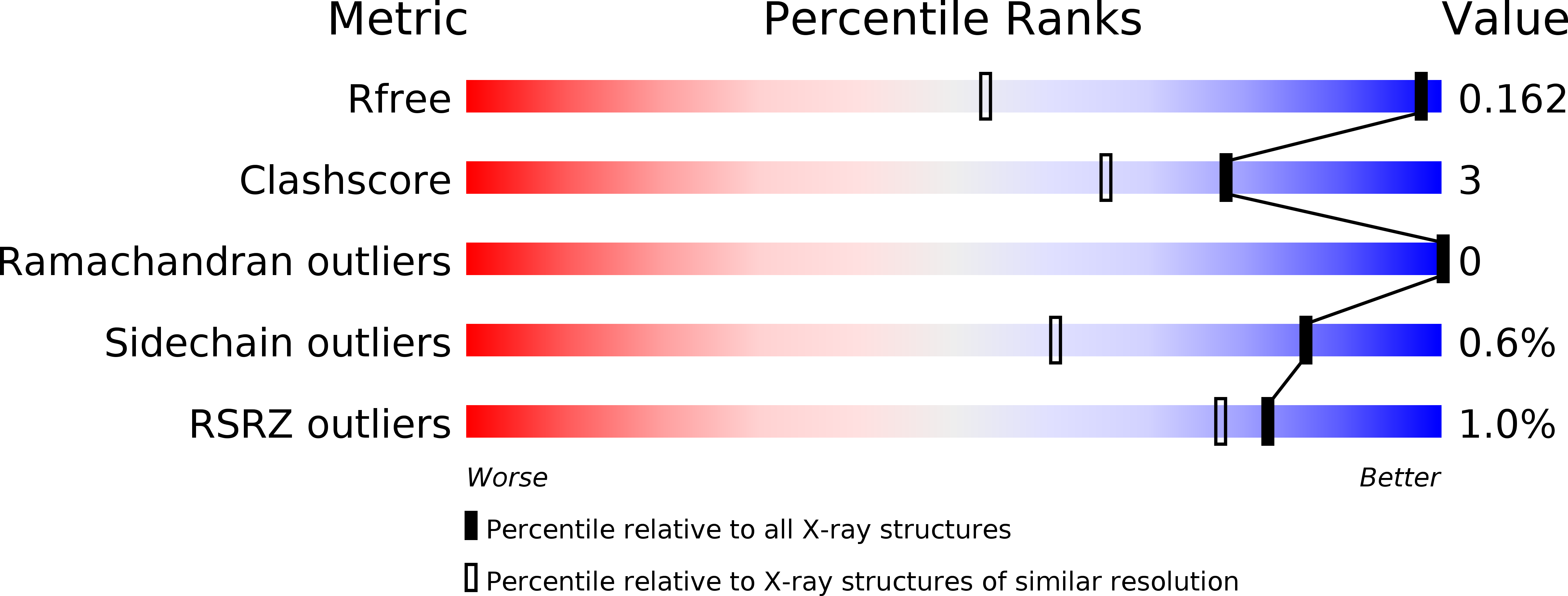
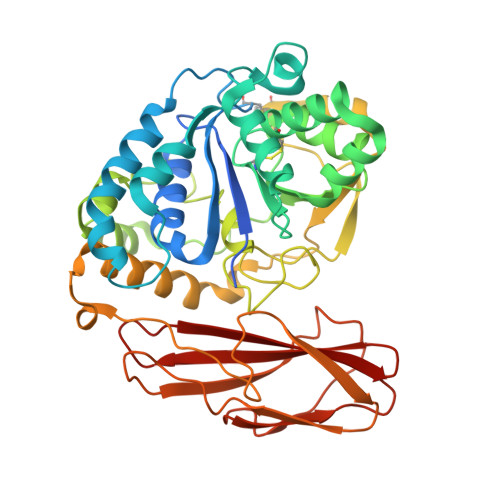
Ab initio phasing of a nucleoside hydrolase-related hypothetical protein from Saccharophagus degradans that is associated with carbohydrate metabolism.
Hehemann, J.H., Marsters, C., Boraston, A.B.(2011) Proteins 79: 2992-2998
- PubMed: 21905122
- DOI: https://doi.org/10.1002/prot.23126
- Primary Citation of Related Structures:
2YHG
Organizational Affiliation:
Biochemistry & Microbiology, University of Victoria, Victoria, BC, Canada.