Structural and Functional Studies on the N-Terminal Domain of the Shigella Type III Secretion Protein Mxig.
Mcdowell, M.A., Johnson, S., Deane, J.E., Cheung, M., Roehrich, A.D., Blocker, A.J., Mcdonnell, J.M., Lea, S.M.(2011) J Biol Chem 286: 30606
- PubMed: 21733840
- DOI: https://doi.org/10.1074/jbc.M111.243865
- Primary Citation of Related Structures:
2XXS - PubMed Abstract:
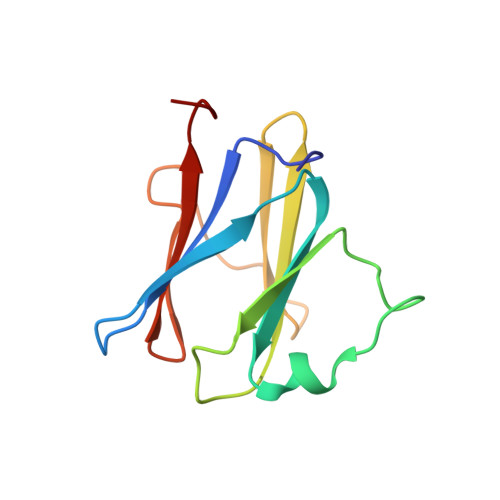
MxiG is a single-pass membrane protein that oligomerizes within the inner membrane ring of the Shigella flexneri type III secretion system (T3SS). The MxiG N-terminal domain (MxiG-N) is the predominant cytoplasmic structure; however, its role in T3SS assembly and secretion is largely uncharacterized. We have determined the solution structure of MxiG-N residues 6-112 (MxiG-N(6-112)), representing the first published structure of this T3SS domain. The structure shows strong structural homology to forkhead-associated (FHA) domains. Canonically, these cell-signaling modules bind phosphothreonine (Thr(P)) via highly conserved residues. However, the putative phosphate-binding pocket of MxiG-N(6-112) does not align with other FHA domain structures or interact with Thr(P). Furthermore, mutagenesis of potential phosphate-binding residues has no effect on S. flexneri T3SS assembly and function. Therefore, MxiG-N has a novel function for an FHA domain. Positioning of MxiG-N(6-112) within the EM density of the S. flexneri needle complex gives insight into the ambiguous stoichiometry of the T3SS, supporting models with 24 MxiG subunits in the inner membrane ring.
Organizational Affiliation:
Sir William Dunn School of Pathology, University of Oxford, Oxford OX1 3RE, United Kingdom.