Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
Chorell, E., Pinkner, J.S., Phan, G., Edvinsson, S., Buelens, F., Remaut, H., Waksman, G., Hultgren, S.J., Almqvist, F.(2010) J Med Chem 53: 5690
- PubMed: 20586493
- DOI: https://doi.org/10.1021/jm100470t
- Primary Citation of Related Structures:
2XG4, 2XG5 - PubMed Abstract:
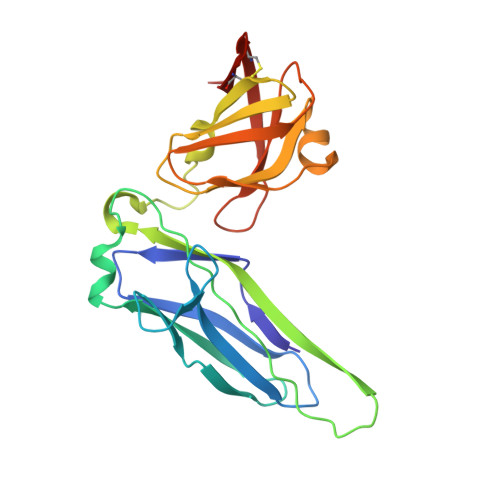
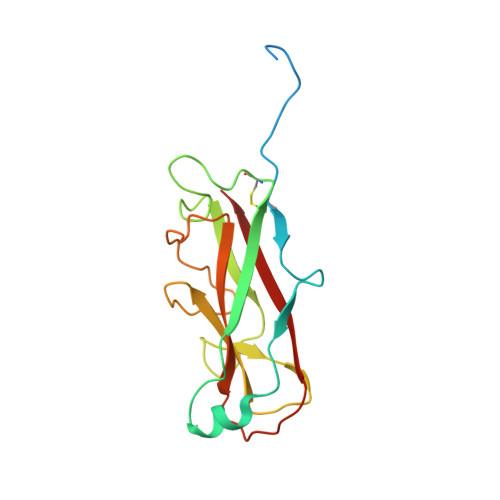
Pilicides block pili formation by binding to pilus chaperones and blocking their function in the chaperone/usher pathway in E. coli. Various C-2 substituents were introduced on the pilicide scaffold by design and synthetic method developments. Experimental evaluation showed that proper substitution of this position affected the biological activity of the compound. Aryl substituents resulted in pilicides with significantly increased potencies as measured in pili-dependent biofilm and hemagglutination assays. The structural basis of the PapD chaperone-pilicide interactions was determined by X-ray crystallography.
Organizational Affiliation:
Department of Chemistry, Umeå University, SE-90187 Umeå, Sweden.