A Protein Encoded by a New Family of Mobile Elements from Euryarchaea Exhibits Three Domains with Novel Folds.
Keller, J., Leulliot, N., Soler, N., Collinet, B., Vincentelli, R., Forterre, P., Van Tilbeurgh, H.(2009) Protein Sci 18: 850
- PubMed: 19319959
- DOI: https://doi.org/10.1002/pro.73
- Primary Citation of Related Structures:
2WB7 - PubMed Abstract:
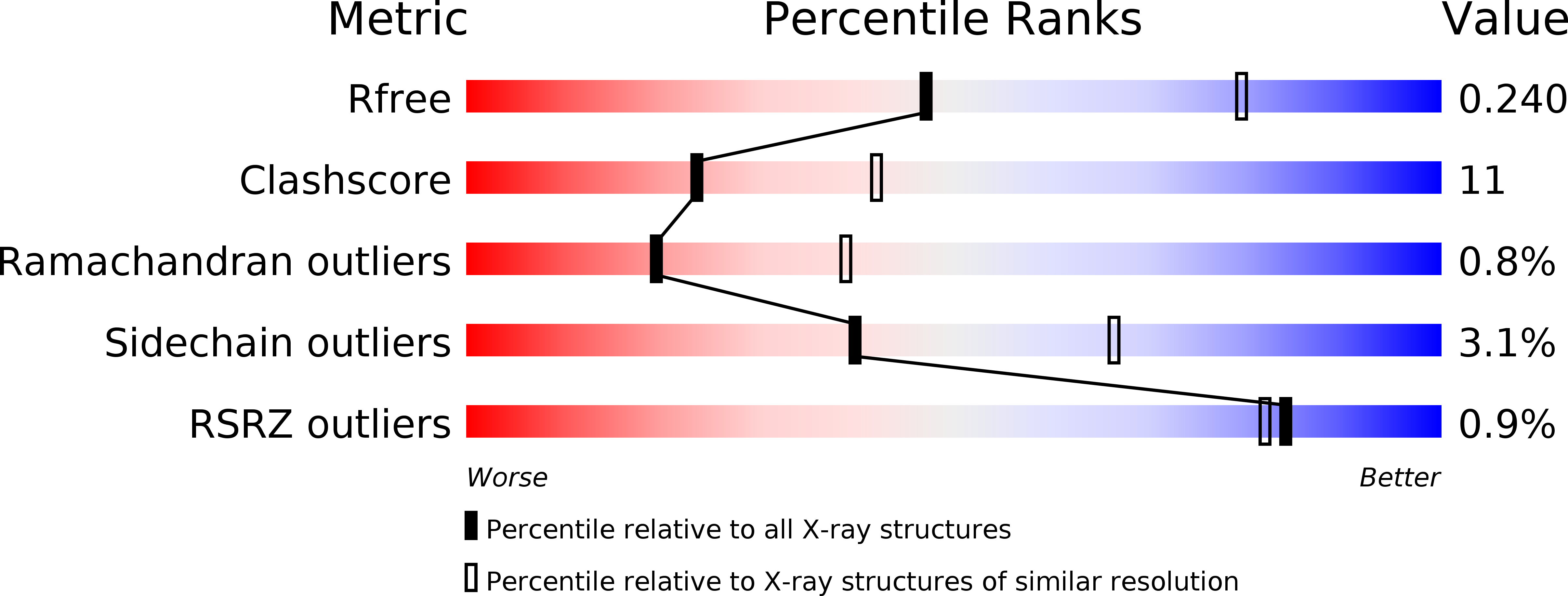
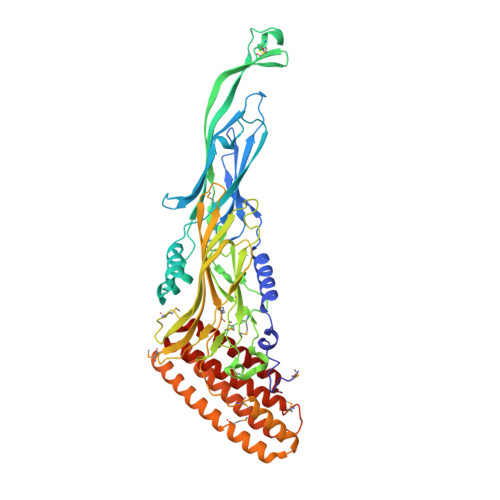
We present here the 2.6A resolution crystal structure of the pT26-6p protein, which is encoded by an ORF of the plasmid pT26-2, recently isolated from the hyperthermophilic archaeon, Thermococcus sp. 26,2. This large protein is present in all members of a new family of mobile elements that, beside pT26-2 include several virus-like elements integrated in the genomes of several Thermococcales and Methanococcales (phylum Euryarchaeota). Phylogenetic analysis suggested that this protein, together with its nearest neighbor (organized as an operon) have coevolved for a long time with the cellular hosts of the encoding mobile element. As the sequences of the N and C-terminal regions suggested a possible membrane association, a deletion construct (739 amino acids) was used for structural analysis. The structure consists of two very similar beta-sheet domains with a new topology and a five helical bundle C-terminal domain. Each of these domains corresponds to a unique fold that has presently not been found in cellular proteins. This result supports the idea that proteins encoded by plasmid and viruses that have no cellular homologues could be a reservoir of new folds for structural genomic studies.
Organizational Affiliation:
Institut de Biochimie et de Biophysique Moléculaire et Cellulaire, Université Paris-Sud, IFR115, UMR8619-CNRS, 91405 Orsay, France.