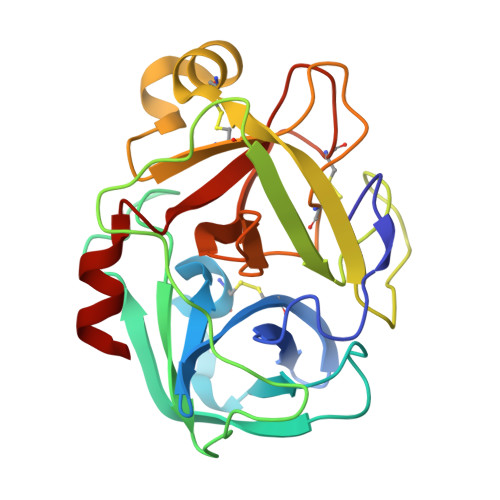
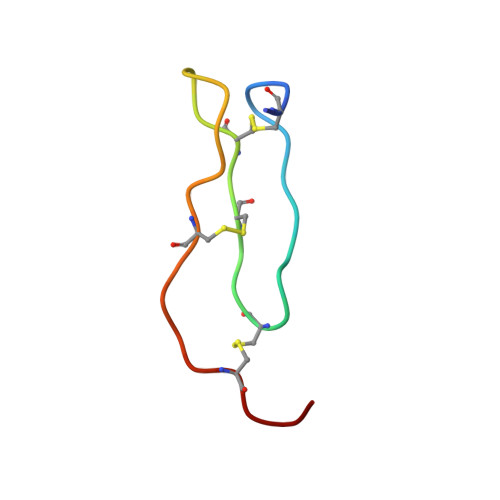
Structure of Locusta Migratoria Protease Inhibitor 3 (Lmpi-3) in Complex with Fusarium Oxysporum Trypsin.
Leone, P., Roussel, A., Kellenberger, C.(2008) Acta Crystallogr D Biol Crystallogr 64: 1165
- PubMed: 19020355
- DOI: https://doi.org/10.1107/S0907444908030400
- Primary Citation of Related Structures:
2VU8 - PubMed Abstract:
Previous studies have shown that the trypsin inhibitors LMPI-1, LMPI-3 and SGTI from locusts display an unusual species selectivity. They inhibit locust, crayfish and fungal trypsins several orders of magnitude more efficiently than bovine trypsin. In contrast, the chymotrypsin inhibitors from the same family, LMPI-2 and SGCI, are active towards mammalian enzymes. The crystal structures of a variant of LMPI-1 and of LMPI-2 in complex with bovine chymotrypsin have revealed subtle structural differences between the trypsin and chymotrypsin inhibitors. In a previous report, it was proposed that Pro173 of bovine trypsin is responsible for the weak inhibitory activity of LMPI-1 and LMPI-3. A fungal trypsin from Fusarium oxysporum contains Gly173 instead of Pro173 and has been shown to be strongly inhibited by LMPI-1 and LMPI-3. To explore the structural features that are responsible for this property, the crystal structure of the complex between LMPI-3 and F. oxysporum trypsin was determined at 1.8 A resolution. This study indicates that this small inhibitor interacts with the protease through the reactive site P3-P4' and the P10-P6 residues. Comparison of this complex with the SGTI-crayfish trypsin and BPTI-bovine trypsin complexes reinforces this hypothesis on the role of residue 173 of trypsin in species selectivity.
Organizational Affiliation:
AFMB UMR6098 Case 932, 163 Avenue de Luminy, 13288 Marseille CEDEX 9, France.