The First Structure of a Glycoside Hydrolase Family 61 Member, Cel61B from the Hypocrea Jecorina, at 1.6 A Resolution.
Karkehabadi, S., Hansson, H., Kim, S., Piens, K., Mitchinson, C., Sandgren, M.(2008) J Mol Biol 383: 144
- PubMed: 18723026
- DOI: https://doi.org/10.1016/j.jmb.2008.08.016
- Primary Citation of Related Structures:
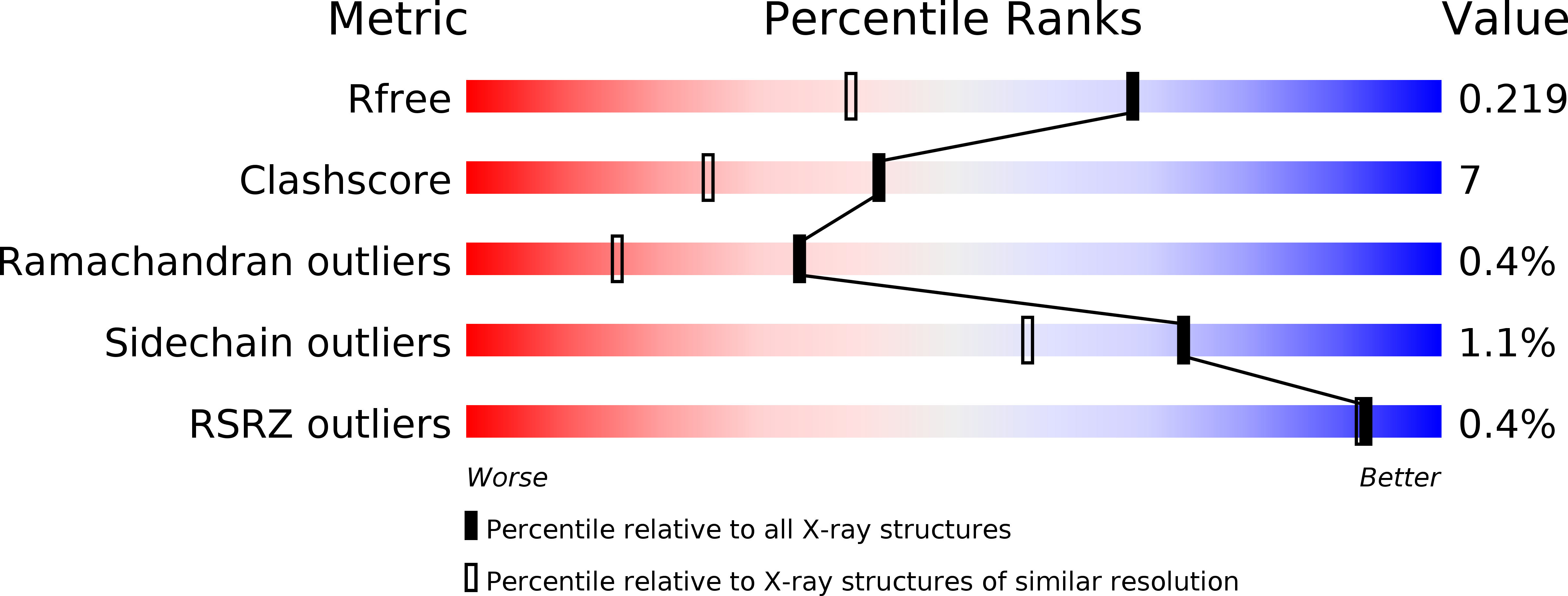
2VTC - PubMed Abstract:
The glycoside hydrolase (GH) family 61 is a long-recognized, but still recondite, class of proteins, with little known about the activity, mechanism or function of its more than 70 members. The best-studied GH family 61 member, Cel61A of the filamentous fungus Hypocrea jecorina, is known to be an endoglucanase, but it is not clear if this represents the main activity or function of this family in vivo. We present here the first structure for this family, that of Cel61B from H. jecorina. The best-quality crystals were formed in the presence of nickel, and the crystal structure was solved to 1.6 A resolution using a single-wavelength anomalous dispersion method with nickel as the source of anomalous scatter. Cel61B lacks a carbohydrate-binding module and is a single-domain protein that folds into a twisted beta-sandwich. A structure-aided sequence alignment of all GH family 61 proteins identified a highly conserved group of residues on the surface of Cel61B. Within this patch of mostly polar amino acids was a site occupied by the intramolecular nickel hexacoordinately bound in the solved structure. In the Cel61B structure, there is no easily identifiable carbohydrate-binding cleft or pocket or catalytic center of the types normally seen in GHs. A structural comparison search showed that the known structure most similar to Cel61B is that of CBP21 from the Gram-negative soil bacterium Serratia marcescens, a member of the carbohydrate-binding module family 33 proteins. A polar surface patch highly conserved in that structural family has been identified in CBP21 and shown to be involved in chitin binding and in the protein's enhancement of chitinase activities. The analysis of the Cel61B structure is discussed in light of our continuing research to better understand the activities and function of GH family 61.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Biomedical Center, Uppsala, Sweden.