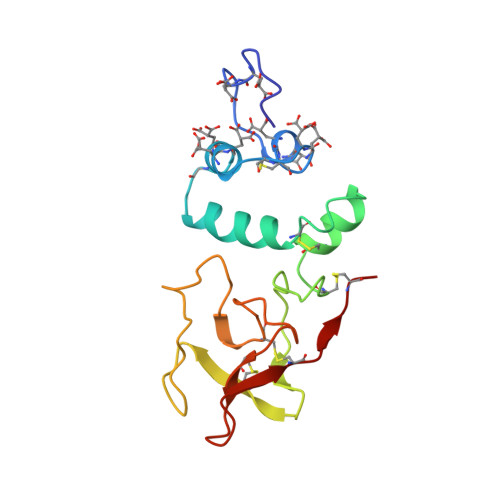
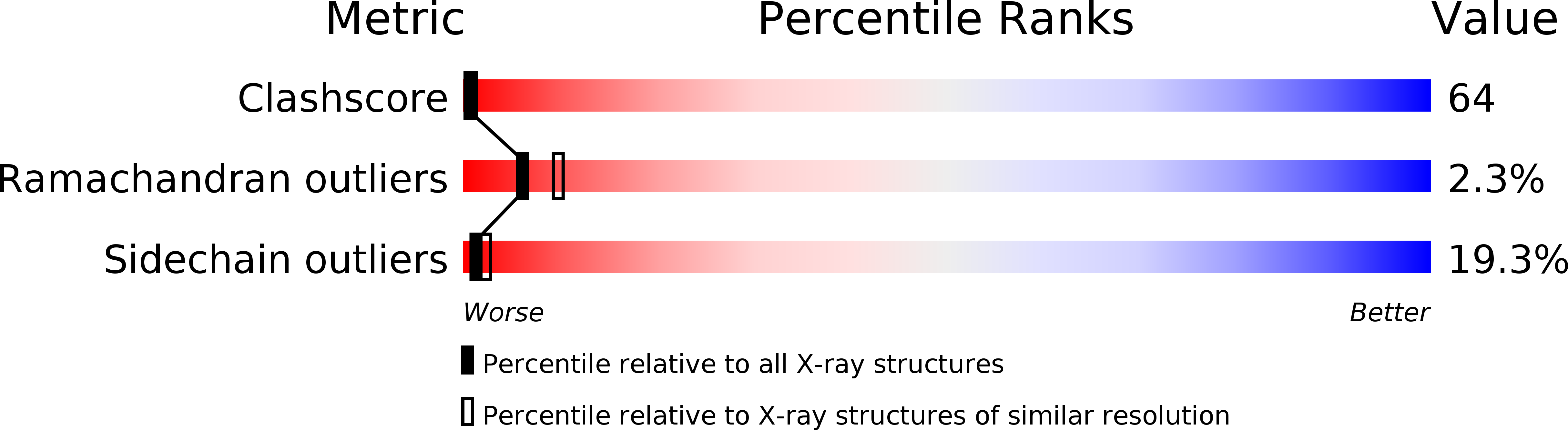Differences in the metal ion structure between Sr- and Ca-prothrombin fragment 1.
Seshadri, T.P., Skrzypczak-Jankun, E., Yin, M., Tulinsky, A.(1994) Biochemistry 33: 1087-1092
- PubMed: 8110739
- DOI: https://doi.org/10.1021/bi00171a006
- Primary Citation of Related Structures:
2SPT - PubMed Abstract:
The structure of Sr-prothrombin fragment 1 has been solved and refined by restrained least-squares methods at 2.5-A resolution to a crystallographic R value of 0.167. The protein structure is very similar to that of Ca-fragment 1. A polymeric array of five Sr2+ ions separated by about 4.0 A is buried among six gamma-carboxyglutamic acid (Gla) residues; three other Sr2+ ions interact with other Gla residues and are located further apart. One of these was not found in the Ca-fragment 1 structure. The coordination of the Sr2+ ions resembles that of Ca2+, but there are some significant differences between them. The most notable is the lack of water coordination with Sr2+ ions and two conformations for Gla 8, which change the coordination of Sr-2 and Sr-3. A hexose moiety of an oligosaccharide was located in the vicinity of Asn101 that was flexibly disordered in Ca-fragment 1. The new Sr2+ ion found may be involved in metal ion phospholipid binding interactions along with Sr-1, and Sr-7, Sr-8.
Organizational Affiliation:
Department of Chemistry, Michigan State University, East Lansing 48824.