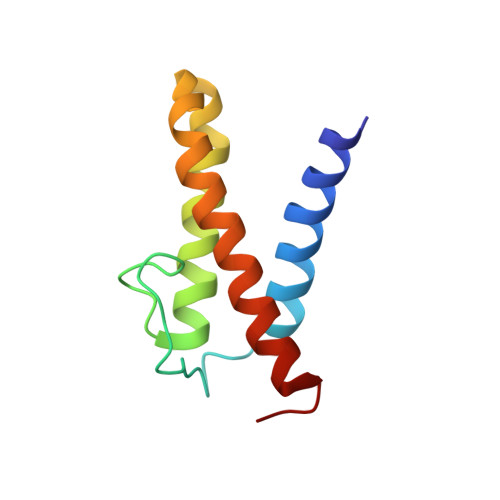
Solution structure of the N-terminal domain of a replication restart primosome factor, PriC, in Escherichia coli.
Aramaki, T., Abe, Y., Katayama, T., Ueda, T.(2013) Protein Sci 22: 1279-1286
- PubMed: 23868391
- DOI: https://doi.org/10.1002/pro.2314
- Primary Citation of Related Structures:
2RT6 - PubMed Abstract:
In eubacterial organisms, the oriC-independent primosome plays an essential role in replication restart after the dissociation of the replication DNA-protein complex by DNA damage. PriC is a key protein component in the replication restart primosome. Our recent study suggested that PriC is divided into two domains: an N-terminal and a C-terminal domain. In the present study, we determined the solution structure of the N-terminal domain, whose structure and function have remained unknown until now. The revealed structure was composed of three helices and one extended loop. We also observed chemical shift changes in the heteronuclear NMR spectrum and oligomerization in the presence of ssDNA. These abilities may contribute to the PriC-ssDNA complex, which is important for the replication restart primosome.
Organizational Affiliation:
Department of Protein Structure, Function and Design, Graduate School of Pharmaceutical Sciences, Kyushu University, Fukuoka, 812-8582, Japan.