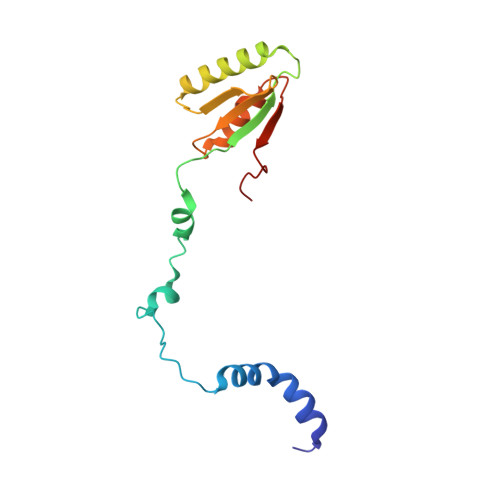
The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Kohler, C., Lighthouse, J.K., Werther, T., Andersen, O.M., Diehl, A., Schmieder, P., Du, J., Holdener, B.C., Oschkinat, H.(2011) Structure 19: 337-348
- PubMed: 21397185
- DOI: https://doi.org/10.1016/j.str.2010.12.022
- Primary Citation of Related Structures:
2RQK, 2RQM - PubMed Abstract:
Mesoderm development (MESD) is a 224 amino acid mouse protein that acts as a molecular chaperone for the low-density lipoprotein receptor (LDLR) family. Here, we provide evidence that the region 45-184 of MESD is essential and sufficient for this function and suggest a model for its mode of action. NMR studies reveal a β-α-β-β-α-β core domain with an α-helical N-terminal extension that interacts with the β sheet in a dynamic manner. As a result, the structural ensemble contains open (active) and closed (inactive) forms, allowing for regulation of chaperone activity through substrate binding. The mutant W61R, which is lethal in Drosophila, adopts only the open state. The receptor motif recognized by MESD was identified by in vitro-binding studies. Furthermore, in vivo functional evidence for the relevance of the identified contact sites in MESD is provided.
Organizational Affiliation:
Department of NMR-Supported Structural Biology, Leibniz-Institut für Molekulare Pharmakologie, Robert-Rössle-Str. 10, 13125 Berlin, Germany.