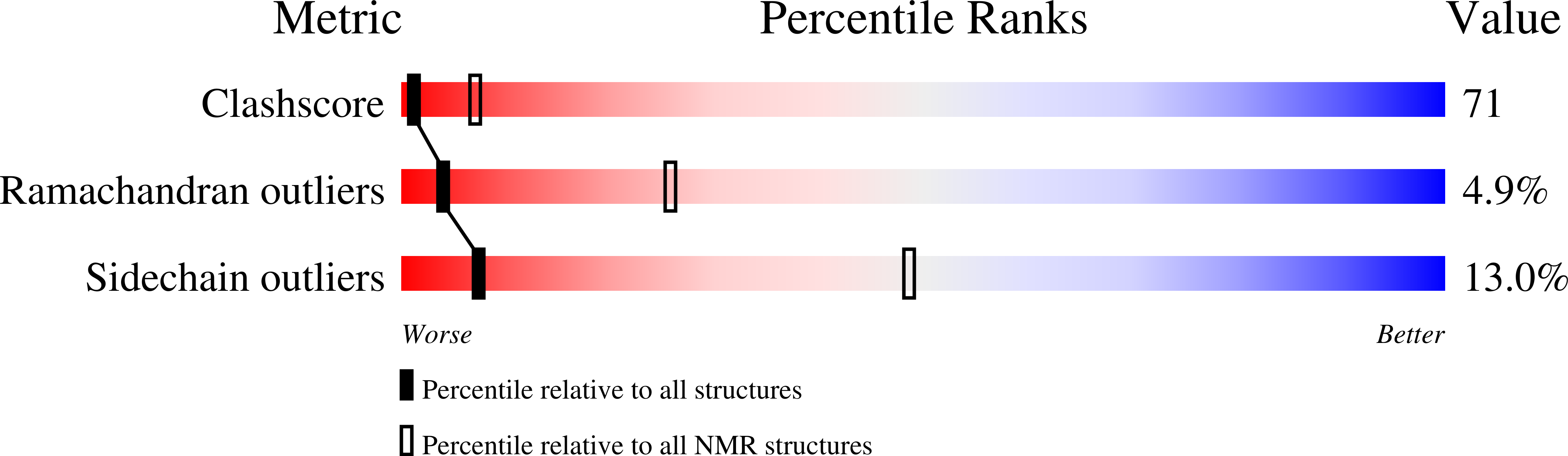Structural basis for a distinct catalytic mechanism in Trypanosoma brucei tryparedoxin peroxidase
Melchers, J., Diechtierow, M., Feher, K., Sinning, I., Tews, I., Krauth-Siegel, R.L., Muhle-Goll, C.(2008) J Biol Chem 283: 30401-30411
- PubMed: 18684708
- DOI: https://doi.org/10.1074/jbc.M803563200
- Primary Citation of Related Structures:
2RM5, 2RM6, 3DWV - PubMed Abstract:
Trypanosoma brucei, the causative agent of African sleeping sickness, encodes three cysteine homologues (Px I-III) of classical selenocysteine-containing glutathione peroxidases. The enzymes obtain their reducing equivalents from the unique trypanothione (bis(glutathionyl)spermidine)/tryparedoxin system. During catalysis, these tryparedoxin peroxidases cycle between an oxidized form with an intramolecular disulfide bond between Cys(47) and Cys(95) and the reduced peroxidase with both residues in the thiol state. Here we report on the three-dimensional structures of oxidized T. brucei Px III at 1.4A resolution obtained by x-ray crystallography and of both the oxidized and the reduced protein determined by NMR spectroscopy. Px III is a monomeric protein unlike the homologous poplar thioredoxin peroxidase (TxP). The structures of oxidized and reduced Px III are essentially identical in contrast to what was recently found for TxP. In Px III, Cys(47), Gln(82), and Trp(137) do not form the catalytic triad observed in the selenoenzymes, and related proteins and the latter two residues are unaffected by the redox state of the protein. The mutational analysis of three conserved lysine residues in the vicinity of the catalytic cysteines revealed that exchange of Lys(107) against glutamate abrogates the reduction of hydrogen peroxide, whereas Lys(97) and Lys(99) play a crucial role in the interaction with tryparedoxin.
Organizational Affiliation:
Department of Structure and Biocomputing, EMBL, 69117 Heidelberg, Germany.