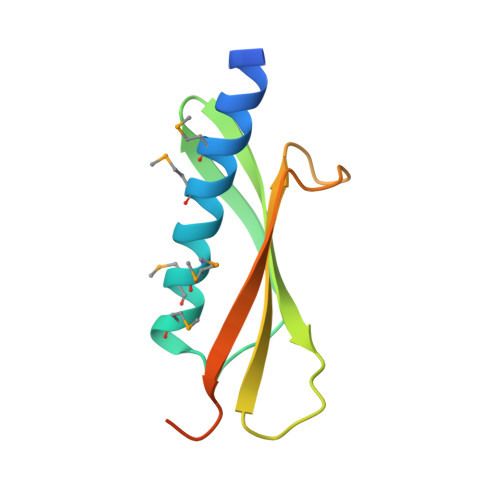
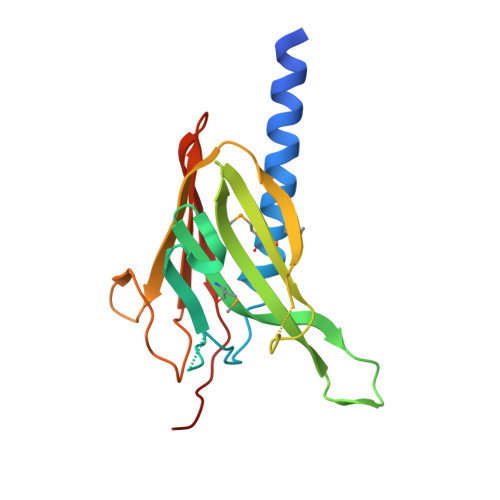
The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus.
Yanez, M.E., Korotkov, K.V., Abendroth, J., Hol, W.G.(2008) J Mol Biol 375: 471-486
- PubMed: 18022192
- DOI: https://doi.org/10.1016/j.jmb.2007.10.035
- Primary Citation of Related Structures:
2RET - PubMed Abstract:
Type II secretion systems (T2SS) translocate virulence factors from the periplasmic space of many pathogenic bacteria into the extracellular environment. The T2SS of Vibrio cholerae and related species is called the extracellular protein secretion (Eps) system that consists of a core of multiple copies of 11 different proteins. The pseudopilins, EpsG, EpsH, EpsI, EpsJ and EpsK, are five T2SS proteins that are thought to assemble into a pseudopilus, which is assumed to interact with the outer membrane pore, and may actively participate in the export of proteins. We report here biochemical evidence that the minor pseudopilins EpsI and EpsJ from Vibrio species interact directly with one another. Moreover, the 2.3 A resolution crystal structure of a complex of EspI and EpsJ from Vibrio vulnificus represents the first atomic resolution structure of a complex of two different pseudopilin components from the T2SS. Both EpsI and EpsJ appear to be structural extremes within the family of type 4a pilin structures solved to date, with EpsI having the smallest, and EpsJ the largest, "variable pilin segment" seen thus far. A high degree of sequence conservation in the EpsI:EpsJ interface indicates that this heterodimer occurs in the T2SS of a large number of bacteria. The arrangement of EpsI and EpsJ in the heterodimer would correspond to a right-handed helical character of proteins assembled into a pseudopilus.
Organizational Affiliation:
Department of Biochemistry, Biomolecular Structure Center, University of Washington, Box 357742, Seattle, WA 98195, USA.