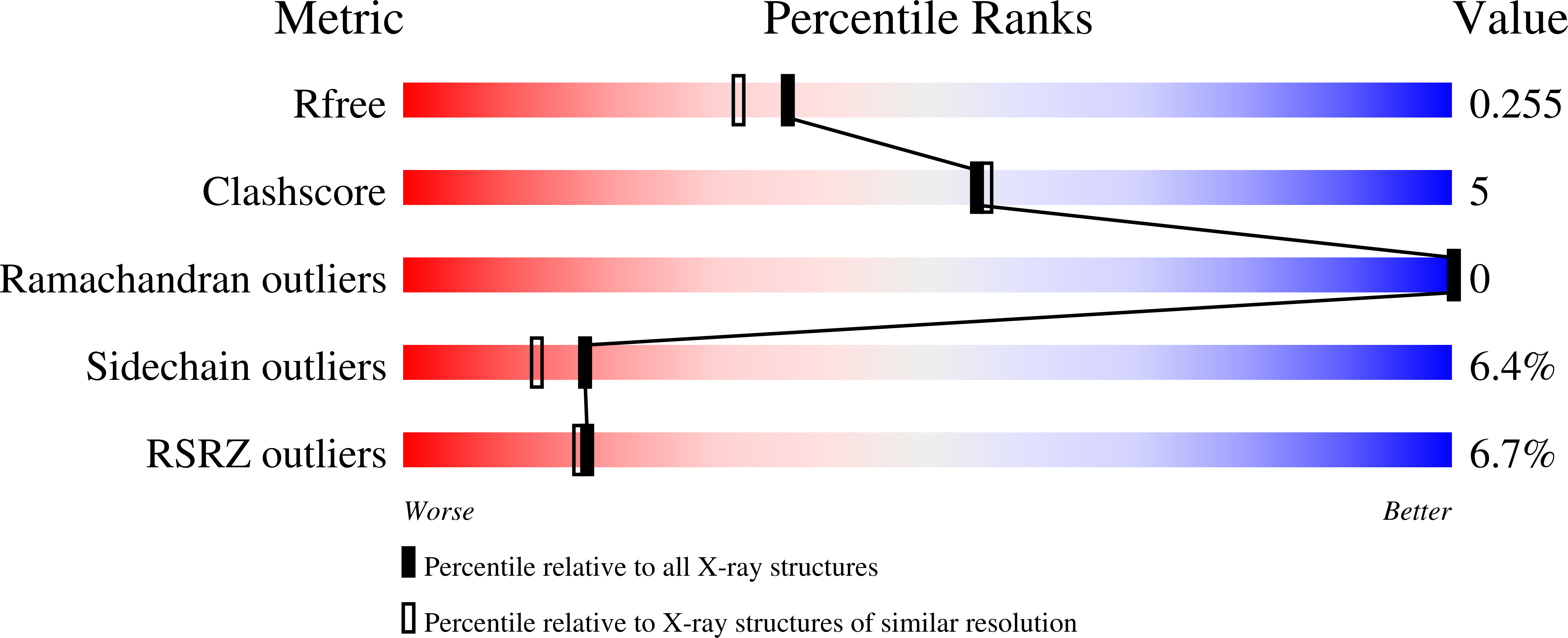
Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum.
Wernimont, A.K., Lew, J., Lin, Y.H., Zhao, Y., Kozieradzki, I., Ravichandran, M., Shapiro, M., Bochkarev, A., Edwards, A.M., Arrowsmith, C.H., Weigelt, J., Sundstrom, M., Hui, R., Qiu, W., Sukumar, D., Hassanali, A.To be published.