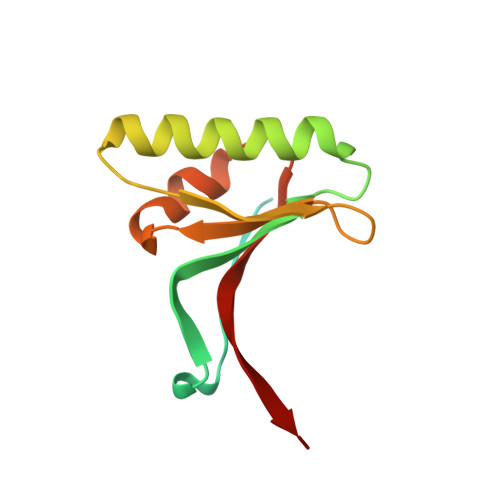
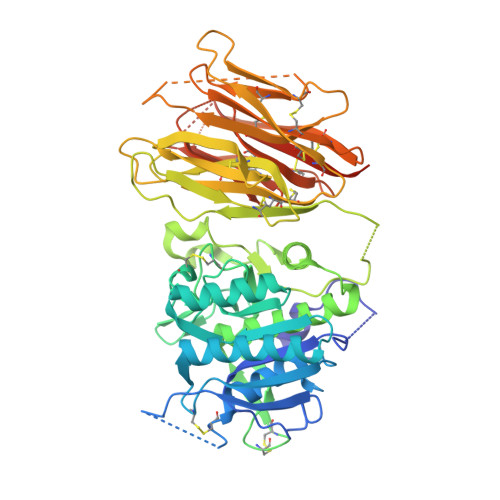
The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Hampton, E.N., Knuth, M.W., Li, J., Harris, J.L., Lesley, S.A., Spraggon, G.(2007) Proc Natl Acad Sci U S A 104: 14604-14609
- PubMed: 17804797
- DOI: https://doi.org/10.1073/pnas.0703402104
- Primary Citation of Related Structures:
2QTW - PubMed Abstract:
Mutations in proprotein convertase subtilisin/kexin type 9 (PCSK9) are strongly associated with levels of low-density lipoprotein cholesterol in the blood plasma and, thereby, occurrence or resistance to atherosclerosis and coronary heart disease. Despite this importance, relatively little is known about the biology of PCSK9. Here, the crystal structure of a full-length construct of PCSK9 solved to 1.9-A resolution is presented. The structure contains a fully folded C-terminal cysteine-rich domain (CRD), showing a distinct structural similarity to the resistin homotrimer, a small cytokine associated with obesity and diabetes. This structural relationship between the CRD of PCSK9 and the resistin family is not observed in primary sequence comparisons and strongly suggests a distant evolutionary link between the two molecules. This three-dimensional homology provides insight into the function of PCSK9 at the molecular level and will help to dissect the link between PCSK9 and CHD.
Organizational Affiliation:
Genomics Institute of the Novartis Research Foundation, 10675 John Jay Hopkins Drive, San Diego, CA 92121, USA.